Analysis of Complete Chloroplast Genome Sequences Improves Phylogenetic Resolution in Paris (Melanthiaceae)
- PMID: 27965698
- PMCID: PMC5126724
- DOI: 10.3389/fpls.2016.01797
Analysis of Complete Chloroplast Genome Sequences Improves Phylogenetic Resolution in Paris (Melanthiaceae)
Abstract
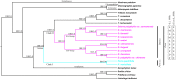
The genus Paris in the broad concept is an economically important group in the monocotyledonous family Melanthiaceae (tribe Parideae). The phylogeny of Paris was controversial in previous morphology-based classification and molecular phylogeny. Here, the complete cp genomes of eleven Paris taxa were sequenced, to better understand the evolutionary relationships among these plants and the mutation patterns in their chloroplast (cp) genomes. Comparative analyses indicated that the overall cp genome structure among the Paris taxa is quite similar. The triplication of trnI-CAU was found only in the cp genomes of P. quadrifolia and P. verticillata. Phylogenetic analyses based on the complete cp genomes did not resolve Paris as a monophyletic group, instead providing evidence supporting division of the twelve taxa into two segregate genera: Paris sensu strict and Daiswa. The sister relationship between Daiswa and Trillium was well supported. We recovered two fully supported lineages with divergent distribution in Daiswa; however, none of the previously recognized sections in Daiswa was resolved as monophyletic using plastome data, suggesting that the infrageneric relationships and biogeography of Daiswa species require further investigation. Ten highly divergent DNA regions, suitable for species identification, were detected among the 12 cp genomes. This study is the first successful attempt to provide well-supported evolutionary relationships in Paris based on phylogenomic analyses. The findings highlight the potential of the whole cp genomes for improving resolution in phylogeny as well as species identification in phylogenetically and taxonomically difficult plant genera.
Keywords: Daiswa; Melanthiaceae; Paris; chloroplast genome; comparative genomics; phylogeny.
Figures



Similar articles
-
Comparative analysis of complete chloroplast genome sequences of five endangered species and new insights into phylogenetic relationships of Paris.Gene. 2022 Jul 30;833:146572. doi: 10.1016/j.gene.2022.146572. Epub 2022 May 21. Gene. 2022. PMID: 35609799
-
A trnI_CAU triplication event in the complete chloroplast genome of Paris verticillata M.Bieb. (Melanthiaceae, Liliales).Genome Biol Evol. 2014 Jun 19;6(7):1699-706. doi: 10.1093/gbe/evu138. Genome Biol Evol. 2014. PMID: 24951560 Free PMC article.
-
Chloroplast phylogenomic analysis provides insights into the evolution of the largest eukaryotic genome holder, Paris japonica (Melanthiaceae).BMC Plant Biol. 2019 Jul 4;19(1):293. doi: 10.1186/s12870-019-1879-7. BMC Plant Biol. 2019. PMID: 31272375 Free PMC article.
-
Insight into infrageneric circumscription through complete chloroplast genome sequences of two Trillium species.AoB Plants. 2016 Apr 6;8:plw015. doi: 10.1093/aobpla/plw015. Print 2016. AoB Plants. 2016. PMID: 26933149 Free PMC article.
-
Research Progress in Plant Molecular Systematics of Lauraceae.Biology (Basel). 2021 May 1;10(5):391. doi: 10.3390/biology10050391. Biology (Basel). 2021. PMID: 34062846 Free PMC article. Review.
Cited by
-
Analysis of complete chloroplast genome sequences and insight into the phylogenetic relationships of Ferula L.BMC Genomics. 2022 Sep 8;23(1):643. doi: 10.1186/s12864-022-08868-z. BMC Genomics. 2022. PMID: 36076164 Free PMC article.
-
Plastid Phylogenomics Provide Evidence to Accept Two New Members of Ligusticopsis (Apiaceae, Angiosperms).Int J Mol Sci. 2022 Dec 26;24(1):382. doi: 10.3390/ijms24010382. Int J Mol Sci. 2022. PMID: 36613825 Free PMC article.
-
Comparative plastome analysis of Musaceae and new insights into phylogenetic relationships.BMC Genomics. 2022 Mar 21;23(1):223. doi: 10.1186/s12864-022-08454-3. BMC Genomics. 2022. PMID: 35313810 Free PMC article.
-
The complete plastome of Panax stipuleanatus: Comparative and phylogenetic analyses of the genus Panax (Araliaceae).Plant Divers. 2018 Nov 22;40(6):265-276. doi: 10.1016/j.pld.2018.11.001. eCollection 2018 Dec. Plant Divers. 2018. PMID: 30740573 Free PMC article.
-
Chloroplast Genomic Resource of Paris for Species Discrimination.Sci Rep. 2017 Jun 13;7(1):3427. doi: 10.1038/s41598-017-02083-7. Sci Rep. 2017. PMID: 28611359 Free PMC article.
References
-
- Angiosperm Phylogeny Group (2016). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 181 1–20. 10.1016/j.jep.2015.05.035 - DOI
-
- Bodin S. S., Kim J. S., Kim J. H. (2013). Complete chloroplast genome of Chionographis japonica (Willd.) Maxim. (Melanthiaceae): comparative genomics and evaluation of universal primers for Liliales. Plant Mol. Biol. Rep. 31 1407–1421. 10.1007/s11105-013-0616-x - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

