Genome-Wide Association Study for Plant Height and Grain Yield in Rice under Contrasting Moisture Regimes
- PMID: 27965699
- PMCID: PMC5126757
- DOI: 10.3389/fpls.2016.01801
Genome-Wide Association Study for Plant Height and Grain Yield in Rice under Contrasting Moisture Regimes
Abstract
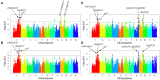
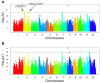
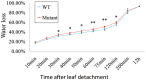
Drought is one of the vitally critical environmental stresses affecting both growth and yield potential in rice. Drought resistance is a complicated quantitative trait that is regulated by numerous small effect loci and hundreds of genes controlling various morphological and physiological responses to drought. For this study, 270 rice landraces and cultivars were analyzed for their drought resistance. This was done via determination of changes in plant height and grain yield under contrasting water regimes, followed by detailed identification of the underlying genetic architecture via genome-wide association study (GWAS). We controlled population structure by setting top two eigenvectors and combining kinship matrix for GWAS in this study. Eighteen, five, and six associated loci were identified for plant height, grain yield per plant, and drought resistant coefficient, respectively. Nine known functional genes were identified, including five for plant height (OsGA2ox3, OsGH3-2, sd-1, OsGNA1, and OsSAP11/OsDOG), two for grain yield per plant (OsCYP51G3 and OsRRMh) and two for drought resistant coefficient (OsPYL2 and OsGA2ox9), implying very reliable results. A previous study reported OsGNA1 to regulate root development, but this study reports additional controlling of both plant height and root length. Moreover, OsRLK5 is a new drought resistant candidate gene discovered in this study. OsRLK5 mutants showed faster water loss rates in detached leaves. This gene plays an important role in the positive regulation of yield-related traits under drought conditions. We furthermore discovered several new loci contributing to the three investigated traits (plant height, grain yield, and drought resistance). These associated loci and candidate genes significantly improve our knowledge of the genetic control of these traits in rice. In addition, many drought resistant cultivars screened in this study can be used as parental genotypes to improve drought resistance of rice by molecular breeding.
Keywords: GWAS; candidate genes; drought resistance; grain yield; plant height.
Figures





Similar articles
-
QTL mapping and analysis for drought tolerance in rice by genome-wide association study.Front Plant Sci. 2023 Jul 25;14:1223782. doi: 10.3389/fpls.2023.1223782. eCollection 2023. Front Plant Sci. 2023. PMID: 37560028 Free PMC article.
-
Variation of drought resistance of rice genotypes released in different years in China.J Sci Food Agric. 2019 Jul;99(9):4430-4438. doi: 10.1002/jsfa.9678. Epub 2019 Apr 19. J Sci Food Agric. 2019. PMID: 30859570
-
Genome-wide Association Study of a Panel of Vietnamese Rice Landraces Reveals New QTLs for Tolerance to Water Deficit During the Vegetative Phase.Rice (N Y). 2019 Jan 28;12(1):4. doi: 10.1186/s12284-018-0258-6. Rice (N Y). 2019. PMID: 30701393 Free PMC article.
-
Genomics-based precision breeding approaches to improve drought tolerance in rice.Biotechnol Adv. 2013 Dec;31(8):1308-18. doi: 10.1016/j.biotechadv.2013.05.004. Epub 2013 May 20. Biotechnol Adv. 2013. PMID: 23702083 Review.
-
MicroRNAs meet with quantitative trait loci: Small powerful players in regulating quantitative yield traits in rice.Wiley Interdiscip Rev RNA. 2019 Nov;10(6):e1556. doi: 10.1002/wrna.1556. Epub 2019 Mar 24. Wiley Interdiscip Rev RNA. 2019. PMID: 31207122 Review.
Cited by
-
Whole-Genome Sequencing and RNA-Seq Reveal Differences in Genetic Mechanism for Flowering Response between Weedy Rice and Cultivated Rice.Int J Mol Sci. 2022 Jan 30;23(3):1608. doi: 10.3390/ijms23031608. Int J Mol Sci. 2022. PMID: 35163531 Free PMC article.
-
A Genome-Wide Association Study Reveals Candidate Genes Related to Salt Tolerance in Rice (Oryza sativa) at the Germination Stage.Int J Mol Sci. 2018 Oct 12;19(10):3145. doi: 10.3390/ijms19103145. Int J Mol Sci. 2018. PMID: 30322083 Free PMC article.
-
GWAS analysis to elucidate genetic composition underlying a photoperiod-insensitive rice population, North Korea.Front Genet. 2022 Dec 7;13:1036747. doi: 10.3389/fgene.2022.1036747. eCollection 2022. Front Genet. 2022. PMID: 36568369 Free PMC article.
-
Drought's physiological footprint: implications for crop improvement in rice.Mol Biol Rep. 2025 Mar 10;52(1):298. doi: 10.1007/s11033-025-10405-6. Mol Biol Rep. 2025. PMID: 40063283 Review.
-
Biotechnological Advances to Improve Abiotic Stress Tolerance in Crops.Int J Mol Sci. 2022 Oct 10;23(19):12053. doi: 10.3390/ijms231912053. Int J Mol Sci. 2022. PMID: 36233352 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

