Discovery and characterization of an F420-dependent glucose-6-phosphate dehydrogenase (Rh-FGD1) from Rhodococcus jostii RHA1
- PMID: 27966048
- PMCID: PMC5352752
- DOI: 10.1007/s00253-016-8038-y
Discovery and characterization of an F420-dependent glucose-6-phosphate dehydrogenase (Rh-FGD1) from Rhodococcus jostii RHA1
Abstract
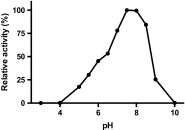
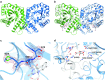
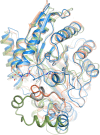
Cofactor F420, a 5-deazaflavin involved in obligatory hydride transfer, is widely distributed among archaeal methanogens and actinomycetes. Owing to the low redox potential of the cofactor, F420-dependent enzymes play a pivotal role in central catabolic pathways and xenobiotic degradation processes in these organisms. A physiologically essential deazaflavoenzyme is the F420-dependent glucose-6-phosphate dehydrogenase (FGD), which catalyzes the reaction F420 + glucose-6-phosphate → F420H2 + 6-phospho-gluconolactone. Thereby, FGDs generate the reduced F420 cofactor required for numerous F420H2-dependent reductases, involved e.g., in the bioreductive activation of the antitubercular prodrugs pretomanid and delamanid. We report here the identification, production, and characterization of three FGDs from Rhodococcus jostii RHA1 (Rh-FGDs), being the first experimental evidence of F420-dependent enzymes in this bacterium. The crystal structure of Rh-FGD1 has also been determined at 1.5 Å resolution, showing a high similarity with FGD from Mycobacterium tuberculosis (Mtb) (Mtb-FGD1). The cofactor-binding pocket and active-site catalytic residues are largely conserved in Rh-FGD1 compared with Mtb-FGD1, except for an extremely flexible insertion region capping the active site at the C-terminal end of the TIM-barrel, which also markedly differs from other structurally related proteins. The role of the three positively charged residues (Lys197, Lys258, and Arg282) constituting the binding site of the substrate phosphate moiety was experimentally corroborated by means of mutagenesis study. The biochemical and structural data presented here provide the first step towards tailoring Rh-FGD1 into a more economical biocatalyst, e.g., an F420-dependent glucose dehydrogenase that requires a cheaper cosubstrate and can better match the demands for the growing applications of F420H2-dependent reductases in industry and bioremediation.
Keywords: Deazaflavoenzymes; F420; Glucose-6-phosphate dehydrogenase; Rhodococcus.
Conflict of interest statement
Funding
This study was funded by a Ubbo Emmius scholarship from the University of Groningen, the Netherlands (awarded to QTN), and the European Community’s Seventh Framework Programme (FP7/2007−2013) under BioStruct-X (Grants 7551 and 10205).
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals by any of the authors.
Figures


References
-
- Ahmed FH, Carr PD, Lee BM, Afriat-Jurnou L, Mohamed AE, Hong N, Flanagan J, Taylor MC, Greening C, Jackson CJ. Sequence–structure–function classification of a catalytically diverse oxidoreductase superfamily in Mycobacteria. J Mol Biol. 2015;427:3554–3571. doi: 10.1016/j.jmb.2015.09.021. - DOI - PubMed
-
- Aufhammer SW, Warkentin E, Ermler U, Hagemeier CH, Thauer RK, Shima S. Crystal structure of methylenetetrahydromethanopterin reductase (Mer) in complex with coenzyme F420: architecture of the F420/FMN binding site of enzymes within the nonprolyl cis-peptide containing bacterial luciferase family. Protein Sci. 2005;14:1840–1849. doi: 10.1110/ps.041289805. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

