New insights into the evolutionary origins of the recombination-activating gene proteins and V(D)J recombination
- PMID: 27973733
- PMCID: PMC5459667
- DOI: 10.1111/febs.13990
New insights into the evolutionary origins of the recombination-activating gene proteins and V(D)J recombination
Abstract
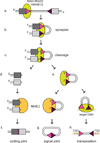
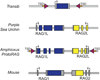
The adaptive immune system of jawed vertebrates relies on V(D)J recombination as one of the main processes to generate the diverse array of receptors necessary for the recognition of a wide range of pathogens. The DNA cleavage reaction necessary for the assembly of the antigen receptor genes from an array of potential gene segments is mediated by the recombination-activating gene proteins RAG1 and RAG2. The RAG proteins have been proposed to originate from a transposable element (TE) as they share mechanistic and structural similarities with several families of transposases and are themselves capable of mediating transposition. A number of RAG-like proteins and TEs with sequence similarity to RAG1 and RAG2 have been identified, but only recently has their function begun to be characterized, revealing mechanistic links to the vertebrate RAGs. Of particular significance is the discovery of ProtoRAG, a transposon superfamily found in the genome of the basal chordate amphioxus. ProtoRAG has many of the sequence and mechanistic features predicted for the ancestral RAG transposon and is likely to be an evolutionary relative of RAG1 and RAG2. In addition, early observations suggesting that RAG1 is able to mediate V(D)J recombination in the absence of RAG2 have been confirmed, implying independent evolutionary origins for the two RAG genes. Here, recent progress in identifying and characterizing RAG-like proteins and the TEs that encode them is summarized and a refined model for the evolution of V(D)J recombination and the RAG proteins is presented.
Keywords: ProtoRAG; RAG-like proteins; RAG1; RAG2; Transib; V(D)J recombination; evolution; transposition.
© 2016 Federation of European Biochemical Societies.
Figures


References
-
- Schatz DG, Oettinger MA, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell. 1989;59:1035–1048. - PubMed
-
- Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. - PubMed
-
- Schatz DG, Swanson PC. V(D)J recombination: mechanisms of initiation. Annu Rev Genet. 2011;45:167–202. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

