Copy Number Heterogeneity of JC Virus Standards
- PMID: 27974546
- PMCID: PMC5328450
- DOI: 10.1128/JCM.02337-16
Copy Number Heterogeneity of JC Virus Standards
Abstract
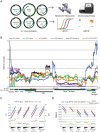
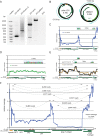
Quantitative PCR is a diagnostic mainstay of clinical virology, and accurate quantitation of viral load among labs requires the use of international standards. However, the use of multiple passages of viral isolates to obtain sufficient material for international standards may result in genomic changes that complicate their use as quantitative standards. We performed next-generation sequencing to obtain single-nucleotide resolution and relative copy number of JC virus (JCV) clinical standards. Strikingly, the WHO international standard and the Exact v1/v2 prototype standards for JCV showed 8-fold and 4-fold variation in genomic coverage between different loci in the viral genome, respectively, due to large deletions in the large T antigen region. Intriguingly, several of the JCV standards sequenced in this study with large T antigen deletions were cultured in cell lines immortalized using simian virus 40 (SV40) T antigen, suggesting the possibility of transcomplementation in cell culture. Using a cutoff 5% allele fraction for junctional reads, 7 different rearrangements were present in the JC virus sequences present in the WHO standard across multiple library preparations and sequencing runs. Neither the copy number differences nor the rearrangements were observed in a clinical sample with a high copy number of JCV or a plasmid control. These results were also confirmed by the quantitative real-time PCR (qPCR), droplet digital PCR (ddPCR), and Sanger sequencing of multiple rearrangements. In summary, targeting different regions of the same international standard can result in up to an 8-fold difference in quantitation. We recommend the use of next-generation sequencing to validate standards in clinical virology.
Keywords: BKV; JC virus; T antigen; clinical standards; deep sequencing; polyomavirus; qPCR; simian virus 40.
Copyright © 2017 Greninger et al.
Figures
Similar articles
-
In Vivo Generation of BK and JC Polyomavirus Defective Viral Genomes in Human Urine Samples Associated with Higher Viral Loads.J Virol. 2021 May 24;95(12):e00250-21. doi: 10.1128/JVI.00250-21. Print 2021 May 24. J Virol. 2021. PMID: 33827948 Free PMC article.
-
Deep Sequencing and Molecular Characterisation of BK Virus and JC Virus WHO International Reference Materials for Clinical Diagnostic Use.Viruses. 2023 May 30;15(6):1289. doi: 10.3390/v15061289. Viruses. 2023. PMID: 37376589 Free PMC article.
-
Specific and quantitative detection of human polyomaviruses BKV and JCV by LightCycler real-time PCR.J Clin Virol. 2006 Jun;36(2):159-62. doi: 10.1016/j.jcv.2006.01.013. Epub 2006 Mar 15. J Clin Virol. 2006. PMID: 16542870
-
Detection of BK virus and JC virus DNA in urine samples from immunocompromised (HIV-infected) and immunocompetent (HIV-non-infected) patients using polymerase chain reaction and microplate hybridisation.J Clin Virol. 2004 Apr;29(4):224-9. doi: 10.1016/S1386-6532(03)00155-0. J Clin Virol. 2004. PMID: 15018849
-
JC virus-associated nephropathy in a renal transplant recipient and comparative analysis of previous cases.Transpl Infect Dis. 2011 Feb;13(1):89-92. doi: 10.1111/j.1399-3062.2010.00567.x. Epub 2010 Sep 6. Transpl Infect Dis. 2011. PMID: 21299772 Review.
Cited by
-
Droplet digital PCR of viral DNA/RNA, current progress, challenges, and future perspectives.J Med Virol. 2021 Jul;93(7):4182-4197. doi: 10.1002/jmv.26846. Epub 2021 Mar 11. J Med Virol. 2021. PMID: 33538349 Free PMC article. Review.
-
Ultrasensitive Capture of Human Herpes Simplex Virus Genomes Directly from Clinical Samples Reveals Extraordinarily Limited Evolution in Cell Culture.mSphere. 2018 Jun 13;3(3):e00283-18. doi: 10.1128/mSphereDirect.00283-18. Print 2018 Jun 27. mSphere. 2018. PMID: 29898986 Free PMC article.
-
In Vivo Generation of BK and JC Polyomavirus Defective Viral Genomes in Human Urine Samples Associated with Higher Viral Loads.J Virol. 2021 May 24;95(12):e00250-21. doi: 10.1128/JVI.00250-21. Print 2021 May 24. J Virol. 2021. PMID: 33827948 Free PMC article.
-
Deep Sequencing and Molecular Characterisation of BK Virus and JC Virus WHO International Reference Materials for Clinical Diagnostic Use.Viruses. 2023 May 30;15(6):1289. doi: 10.3390/v15061289. Viruses. 2023. PMID: 37376589 Free PMC article.
-
Applications of Digital PCR for Clinical Microbiology.J Clin Microbiol. 2017 Jun;55(6):1621-1628. doi: 10.1128/JCM.00211-17. Epub 2017 Mar 15. J Clin Microbiol. 2017. PMID: 28298452 Free PMC article. Review.
References
-
- DeCaprio JA, Imperiale MJ, Major EO. 2013. Polyomaviruses, p 1633–1661. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA.
-
- Yu G, Greninger AL, Isa P, Phan TG, Martínez MA, de la Luz Sanchez M, Contreras JF, Santos-Preciado JI, Parsonnet J, Miller S, DeRisi JL, Delwart E, Arias CF, Chiu CY. 2012. Discovery of a novel polyomavirus in acute diarrheal samples from children. PLoS One 7:e49449. doi: 10.1371/journal.pone.0049449. - DOI - PMC - PubMed
-
- Lim ES, Reyes A, Antonio M, Saha D, Ikumapayi UN, Adeyemi M, Stine OC, Skelton R, Brennan DC, Mkakosya RS, Manary MJ, Gordon JI, Wang D. 2013. Discovery of STL polyomavirus, a polyomavirus of ancestral recombinant origin that encodes a unique T antigen by alternative splicing. Virology 436:295–303. doi: 10.1016/j.virol.2012.12.005. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

