IRAV (FLJ11286), an Interferon-Stimulated Gene with Antiviral Activity against Dengue Virus, Interacts with MOV10
- PMID: 27974568
- PMCID: PMC5309953
- DOI: 10.1128/JVI.01606-16
IRAV (FLJ11286), an Interferon-Stimulated Gene with Antiviral Activity against Dengue Virus, Interacts with MOV10
Abstract
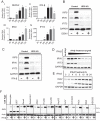
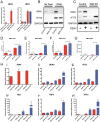
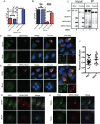
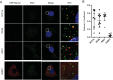
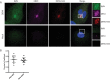
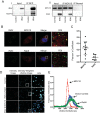
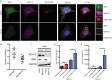
Dengue virus (DENV) is a member of the genus Flavivirus and can cause severe febrile illness. Here, we show that FLJ11286, which we refer to as IRAV, is induced by DENV in an interferon-dependent manner, displays antiviral activity against DENV, and localizes to the DENV replication complex. IRAV is an RNA binding protein and localizes to cytoplasmic processing bodies (P bodies) in uninfected cells, where it interacts with the MOV10 RISC complex RNA helicase, suggesting a role for IRAV in the processing of viral RNA. After DENV infection, IRAV, along with MOV10 and Xrn1, localizes to the DENV replication complex and associates with DENV proteins. Depletion of IRAV or MOV10 results in an increase in viral RNA. These data serve to characterize an interferon-stimulated gene with antiviral activity against DENV, as well as to propose a mechanism of activity involving the processing of viral RNA. IMPORTANCE Dengue virus, a member of the family Flaviviridae, can result in a life-threatening illness and has a significant impact on global health. Dengue virus has been shown to be particularly sensitive to the effects of type I interferon; however, little is known about the mechanisms by which interferon-stimulated genes function to inhibit viral replication. A better understanding of the interferon-mediated antiviral response to dengue virus may aid in the development of novel therapeutics. Here, we examine the influence of the interferon-stimulated gene IRAV (FLJ11286) on dengue virus replication. We show that IRAV associates with P bodies in uninfected cells and with the dengue virus replication complex after infection. IRAV also interacts with MOV10, depletion of which is associated with increased viral replication. Our results provide insight into a newly identified antiviral gene, as well as broadening our understanding of the innate immune response to dengue virus infection.
Keywords: C19orf66; FLJ11286; IRAV; MOV10; P body; UPF0515; dengue; flavivirus; interferon.
Copyright © 2017 Balinsky et al.
Figures

References
-
- Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GRW, Simmons CP, Scott TW, Farrar JJ, Hay SI. 2013. The global distribution and burden of dengue. Nature 496:504–507. doi: 10.1038/nature12060. - DOI - PMC - PubMed
-
- WHO. 2006. WHO scientific working group report on dengue; reference number TDR/SWG/08. WHO, Geneva, Switzerland.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

