The seahorse genome and the evolution of its specialized morphology
- PMID: 27974754
- PMCID: PMC8127814
- DOI: 10.1038/nature20595
The seahorse genome and the evolution of its specialized morphology
Abstract
Seahorses have a specialized morphology that includes a toothless tubular mouth, a body covered with bony plates, a male brood pouch, and the absence of caudal and pelvic fins. Here we report the sequencing and de novo assembly of the genome of the tiger tail seahorse, Hippocampus comes. Comparative genomic analysis identifies higher protein and nucleotide evolutionary rates in H. comes compared with other teleost fish genomes. We identified an astacin metalloprotease gene family that has undergone expansion and is highly expressed in the male brood pouch. We also find that the H. comes genome lacks enamel matrix protein-coding proline/glutamine-rich secretory calcium-binding phosphoprotein genes, which might have led to the loss of mineralized teeth. tbx4, a regulator of hindlimb development, is also not found in H. comes genome. Knockout of tbx4 in zebrafish showed a 'pelvic fin-loss' phenotype similar to that of seahorses.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



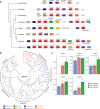







Comment in
-
Sequence reveals genes behind bizarre sea-horse traits.Nature. 2016 Dec 14;540(7633):316. doi: 10.1038/540316a. Nature. 2016. PMID: 27974783 No abstract available.
References
-
- Leysen H, et al. Musculoskeletal structure of the feeding system and implications of snout elongation in Hippocampus reidi and Dunckerocampus dactyliophorus. J. Fish Biol. 2011;78:1799–1823. - PubMed
-
- Stölting KN, Wilson AB. Male pregnancy in seahorses and pipefish: beyond the mammalian model. BioEssays. 2007;29:884–896. - PubMed
-
- Wilson AB, Vincent A, Ahnesjö I, Meyer A. Male pregnancy in seahorses and pipefishes (family Syngnathidae): rapid diversification of paternal brood pouch morphology inferred from a molecular phylogeny. J. Hered. 2001;92:159–166. - PubMed
-
- Teske PR, Cherry MI, Matthee CA. The evolutionary history of seahorses (Syngnathidae: Hippocampus): molecular data suggest a West Pacific origin and two invasions of the Atlantic Ocean. Mol. Phylogenet. Evol. 2004;30:273–286. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

