CanProVar 2.0: An Updated Database of Human Cancer Proteome Variation
- PMID: 27977206
- PMCID: PMC5515284
- DOI: 10.1021/acs.jproteome.6b00505
CanProVar 2.0: An Updated Database of Human Cancer Proteome Variation
Abstract
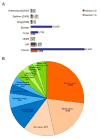
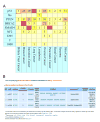
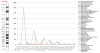
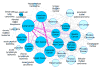
Identification and annotation of the mutations involved in oncogenesis and tumor progression are crucial for both cancer biology and clinical applications. Previously, we developed a public resource CanProVar, a human cancer proteome variation database for storing and querying single amino acid alterations in the human cancers. Since the publication of CanProVar, extensive cancer genomics efforts have revealed the enormous genomic complexity of various types of human cancers. Thus, there is an overwhelming need for comprehensive annotation of the genomic alterations at the protein level and making such knowledge easily accessible. Here, we describe CanProVar 2.0, a significantly expanded version of CanProVar, in which the amount of cancer-related variations and noncancer specific variations was increased by about 10-fold as compared to the previous version. To facilitate the interpretation of the variations, we added to the database functional data on potential impact of the cancer-related variations on 3D protein interaction and on the differential expression of the variant-bearing proteins between cancer and normal samples. The web interface allows for flexible queries based on gene or protein IDs, cancer types, chromosome locations, or pathways. An integrated protein sequence database containing variations that can be directly used for proteomics database searching can be downloaded.
Keywords: cancer; database; proteome; variation.
Figures





Similar articles
-
CanProVar: a human cancer proteome variation database.Hum Mutat. 2010 Mar;31(3):219-28. doi: 10.1002/humu.21176. Hum Mutat. 2010. PMID: 20052754 Free PMC article.
-
dbDEPC 2.0: updated database of differentially expressed proteins in human cancers.Nucleic Acids Res. 2012 Jan;40(Database issue):D964-71. doi: 10.1093/nar/gkr936. Epub 2011 Nov 16. Nucleic Acids Res. 2012. PMID: 22096234 Free PMC article.
-
A bioinformatics workflow for variant peptide detection in shotgun proteomics.Mol Cell Proteomics. 2011 May;10(5):M110.006536. doi: 10.1074/mcp.M110.006536. Epub 2011 Mar 9. Mol Cell Proteomics. 2011. PMID: 21389108 Free PMC article.
-
Proteome informatics for cancer research: from molecules to clinic.Proteomics. 2007 Mar;7(6):976-91. doi: 10.1002/pmic.200600965. Proteomics. 2007. PMID: 17370257 Review.
-
Systematics for types and effects of DNA variations.BMC Genomics. 2018 Dec 28;19(1):974. doi: 10.1186/s12864-018-5262-0. BMC Genomics. 2018. PMID: 30591019 Free PMC article. Review.
Cited by
-
A structurally informed human protein-protein interactome reveals proteome-wide perturbations caused by disease mutations.Nat Biotechnol. 2024 Oct 24. doi: 10.1038/s41587-024-02428-4. Online ahead of print. Nat Biotechnol. 2024. PMID: 39448882
-
MutCombinator: identification of mutated peptides allowing combinatorial mutations using nucleotide-based graph search.Bioinformatics. 2020 Jul 1;36(Suppl_1):i203-i209. doi: 10.1093/bioinformatics/btaa504. Bioinformatics. 2020. PMID: 32657416 Free PMC article.
-
Clinical protein science in translational medicine targeting malignant melanoma.Cell Biol Toxicol. 2019 Aug;35(4):293-332. doi: 10.1007/s10565-019-09468-6. Epub 2019 Mar 21. Cell Biol Toxicol. 2019. PMID: 30900145 Free PMC article.
-
Structurally-informed human interactome reveals proteome-wide perturbations by disease mutations.bioRxiv [Preprint]. 2024 Feb 1:2023.04.24.538110. doi: 10.1101/2023.04.24.538110. bioRxiv. 2024. Update in: Nat Biotechnol. 2024 Oct 24. doi: 10.1038/s41587-024-02428-4. PMID: 37162909 Free PMC article. Updated. Preprint.
-
Single Amino Acid Variant Discovery in Small Numbers of Cells.J Proteome Res. 2019 Jan 4;18(1):417-425. doi: 10.1021/acs.jproteome.8b00694. Epub 2018 Nov 21. J Proteome Res. 2019. PMID: 30404448 Free PMC article.
References
-
- Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153(1):17–37. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

