Transcriptional regulation of core autophagy and lysosomal genes by the androgen receptor promotes prostate cancer progression
- PMID: 27977328
- PMCID: PMC5361609
- DOI: 10.1080/15548627.2016.1268300
Transcriptional regulation of core autophagy and lysosomal genes by the androgen receptor promotes prostate cancer progression
Abstract
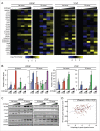
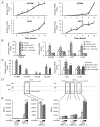
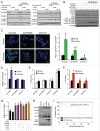
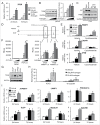
AR (androgen receptor) signaling is crucial for the development and maintenance of the prostate as well as the initiation and progression of prostate cancer. Despite the AR's central role in prostate cancer progression, it is still unclear which AR-mediated processes drive the disease. Here, we identified 4 core autophagy genes: ATG4B, ATG4D, ULK1, and ULK2, in addition to the transcription factor TFEB, a master regulator of lysosomal biogenesis and function, as transcriptional targets of AR in prostate cancer. These findings were significant in light of our recent observation that androgens promoted prostate cancer cell growth in part through the induction of autophagy. Expression of these 5 genes was essential for maximal androgen-mediated autophagy and cell proliferation. In addition, expression of each of these 5 genes alone or in combination was sufficient to increase prostate cancer cell growth independent of AR activity. Further, bioinformatic analysis demonstrated that the expression of these genes correlated with disease progression in 3 separate clinical cohorts. Collectively, these findings demonstrate a functional role for increased autophagy in prostate cancer progression, provide a mechanism for how autophagy is augmented, and highlight the potential of targeting this process for the treatment of advanced prostate cancer.
Keywords: ATG4B; ATG4D; TFEB; ULK1; ULK2; androgen; androgen receptor (AR); autophagy; nuclear receptor; prostate cancer.
Figures

References
-
- Koochekpour S. Androgen receptor signaling and mutations in prostate cancer. Asian J Androl 2010; 12:639-57; PMID:20711217; http://dx.doi.org/10.1038/aja.2010.89 - DOI - PMC - PubMed
-
- Heinlein CA, Chang C. Androgen receptor (AR) coregulators: an overview. Endocr Rev 2002; 23:175-200; PMID:11943742; http://dx.doi.org/10.1210/edrv.23.2.0460 - DOI - PubMed
-
- Knudsen KE, Scher HI. Starving the addiction: new opportunities for durable suppression of AR signaling in prostate cancer. Clin Cancer Res 2009; 15:4792-8; PMID:19638458; http://dx.doi.org/10.1158/1078-0432.CCR-08-2660 - DOI - PMC - PubMed
-
- Shen MM, Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev 2010; 24:1967-2000; PMID:20844012; http://dx.doi.org/10.1101/gad.1965810 - DOI - PMC - PubMed
-
- Society AC. Cancer Facts & Figures 2015. Atlanta: American Cancer Society, 2015.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
