Recruitment Kinetics of Tropomyosin Tpm3.1 to Actin Filament Bundles in the Cytoskeleton Is Independent of Actin Filament Kinetics
- PMID: 27977753
- PMCID: PMC5158027
- DOI: 10.1371/journal.pone.0168203
Recruitment Kinetics of Tropomyosin Tpm3.1 to Actin Filament Bundles in the Cytoskeleton Is Independent of Actin Filament Kinetics
Abstract
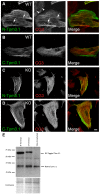
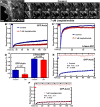
The actin cytoskeleton is a dynamic network of filaments that is involved in virtually every cellular process. Most actin filaments in metazoa exist as a co-polymer of actin and tropomyosin (Tpm) and the function of an actin filament is primarily defined by the specific Tpm isoform associated with it. However, there is little information on the interdependence of these co-polymers during filament assembly and disassembly. We addressed this by investigating the recovery kinetics of fluorescently tagged isoform Tpm3.1 into actin filament bundles using FRAP analysis in cell culture and in vivo in rats using intracellular intravital microscopy, in the presence or absence of the actin-targeting drug jasplakinolide. The mobile fraction of Tpm3.1 is between 50% and 70% depending on whether the tag is at the C- or N-terminus and whether the analysis is in vivo or in cultured cells. We find that the continuous dynamic exchange of Tpm3.1 is not significantly impacted by jasplakinolide, unlike tagged actin. We conclude that tagged Tpm3.1 may be able to undergo exchange in actin filament bundles largely independent of the assembly and turnover of actin.
Conflict of interest statement
PWG is a Director on the Board of Novogen, a company commercialising drugs that are directed against Tpm3.1. This does not alter our adherence to PLOS ONE policies on sharing data and materials.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

