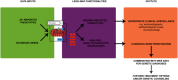
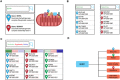
Leigh map: A novel computational diagnostic resource for mitochondrial disease
- PMID: 27977873
- PMCID: PMC5347854
- DOI: 10.1002/ana.24835
Leigh map: A novel computational diagnostic resource for mitochondrial disease
Figures

References
-
- Lightowlers RN, Taylor RW, Turnbull DM. Mutations causing mitochondrial disease: what is new and what challenges remain? Science 2015;349:1494. - PubMed
-
- Lake NJ, Compton AG, Rahman S, et al. Leigh syndrome: one disorder, more than 75 monogenic causes. Ann Neurol 2016;79:190–203. - PubMed
-
- Rahman S, Blok RB, Dahl H‐HM, et al. Leigh syndrome: clinical features and biochemical and DNA abnormalities. Ann Neurol 1996;39:343–351. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

