The mTOR Complex Controls HIV Latency
- PMID: 27978436
- PMCID: PMC5354304
- DOI: 10.1016/j.chom.2016.11.001
The mTOR Complex Controls HIV Latency
Abstract
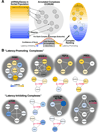
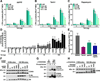
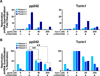
A population of CD4 T lymphocytes harboring latent HIV genomes can persist in patients on antiretroviral therapy, posing a barrier to HIV eradication. To examine cellular complexes controlling HIV latency, we conducted a genome-wide screen with a pooled ultracomplex shRNA library and in vitro system modeling HIV latency and identified the mTOR complex as a modulator of HIV latency. Knockdown of mTOR complex subunits or pharmacological inhibition of mTOR activity suppresses reversal of latency in various HIV-1 latency models and HIV-infected patient cells. mTOR inhibitors suppress HIV transcription both through the viral transactivator Tat and via Tat-independent mechanisms. This inhibition occurs at least in part via blocking the phosphorylation of CDK9, a p-TEFb complex member that serves as a cofactor for Tat-mediated transcription. The control of HIV latency by mTOR signaling identifies a pathway that may have significant therapeutic opportunities.
Keywords: HIV LTR; HIV latency; HIV transcription; genome-wide shRNA screen; latency reversal; mTOR inhibition; reactivation from latency.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

