Cardiovascular proteomics in the era of big data: experimental and computational advances
- PMID: 27980500
- PMCID: PMC5137214
- DOI: 10.1186/s12014-016-9124-y
Cardiovascular proteomics in the era of big data: experimental and computational advances
Abstract
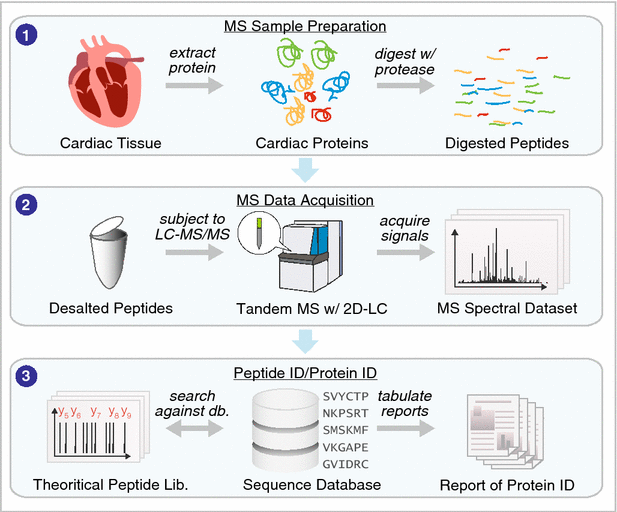
Proteomics plays an increasingly important role in our quest to understand cardiovascular biology. Fueled by analytical and computational advances in the past decade, proteomics applications can now go beyond merely inventorying protein species, and address sophisticated questions on cardiac physiology. The advent of massive mass spectrometry datasets has in turn led to increasing intersection between proteomics and big data science. Here we review new frontiers in technological developments and their applications to cardiovascular medicine. The impact of big data science on cardiovascular proteomics investigations and translation to medicine is highlighted.
Keywords: Cardiovascular medicine; Clinical proteomics; Mass spectrometry; Shotgun proteomics.
Figures



Similar articles
-
Big science and big data in nephrology.Kidney Int. 2019 Jun;95(6):1326-1337. doi: 10.1016/j.kint.2018.11.048. Epub 2019 Mar 5. Kidney Int. 2019. PMID: 30982672 Review.
-
Big Data in Plant Science: Resources and Data Mining Tools for Plant Genomics and Proteomics.Methods Mol Biol. 2016;1415:533-47. doi: 10.1007/978-1-4939-3572-7_27. Methods Mol Biol. 2016. PMID: 27115651
-
Personalized medicine beyond genomics: alternative futures in big data-proteomics, environtome and the social proteome.J Neural Transm (Vienna). 2017 Jan;124(1):25-32. doi: 10.1007/s00702-015-1489-y. Epub 2015 Dec 8. J Neural Transm (Vienna). 2017. PMID: 26645377 Review.
-
New frontiers in proteomics research: a perspective.Int J Pharm. 2005 Aug 11;299(1-2):1-18. doi: 10.1016/j.ijpharm.2005.04.010. Int J Pharm. 2005. PMID: 15979831 Review.
-
Recent developments in proteome informatics for mass spectrometry analysis.Comb Chem High Throughput Screen. 2009 Feb;12(2):194-202. doi: 10.2174/138620709787315508. Comb Chem High Throughput Screen. 2009. PMID: 19199887 Review.
Cited by
-
MiR-146a/b: a family with shared seeds and different roots.Physiol Genomics. 2017 Apr 1;49(4):243-252. doi: 10.1152/physiolgenomics.00133.2016. Epub 2017 Feb 17. Physiol Genomics. 2017. PMID: 28213571 Free PMC article. Review.
-
Possibilities of Proteomics Profiling in Predicting Dysfunction of the Cardiovascular System.Front Physiol. 2022 Apr 25;13:897694. doi: 10.3389/fphys.2022.897694. eCollection 2022. Front Physiol. 2022. PMID: 35547587 Free PMC article. No abstract available.
-
ThermoRawFileParser: Modular, Scalable, and Cross-Platform RAW File Conversion.J Proteome Res. 2020 Jan 3;19(1):537-542. doi: 10.1021/acs.jproteome.9b00328. Epub 2019 Dec 6. J Proteome Res. 2020. PMID: 31755270 Free PMC article.
-
PGCA: An algorithm to link protein groups created from MS/MS data.PLoS One. 2017 May 31;12(5):e0177569. doi: 10.1371/journal.pone.0177569. eCollection 2017. PLoS One. 2017. PMID: 28562641 Free PMC article.
References
-
- Glatter T, Ludwig C, Ahrne E, Aebersold R, Heck AJ, Schmidt A. Large-scale quantitative assessment of different in-solution protein digestion protocols reveals superior cleavage efficiency of tandem Lys-C/trypsin proteolysis over trypsin digestion. J Proteome Res. 2012;11(11):5145–5156. doi: 10.1021/pr300273g. - DOI - PubMed
-
- Lindsey ML, Mayr M, Gomes AV, Delles C, Arrell DK, Murphy AM, Lange RA, Costello CE, Jin Y-F, Laskowitz DT, et al. Transformative impact of proteomics on cardiovascular health and disease: a scientific statement from the American Heart Association. Circulation. 2015;132(9):852–872. doi: 10.1161/CIR.0000000000000226. - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
