Enzymatic incorporation and utilization of an emissive 6-azauridine
- PMID: 27981333
- PMCID: PMC5243813
- DOI: 10.1039/c6ob02080a
Enzymatic incorporation and utilization of an emissive 6-azauridine
Abstract
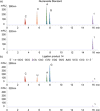
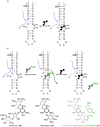
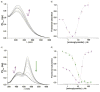
To display favorable fluorescent properties, the non-emissive native nucleosides need to be modified. Here we present a motif that relies on conjugating 5-membered aromatic heterocycles (e.g., thiophene) to a 6-azapyrimidine (1,2,4-triazine) core. Synthetic accessibility and desirable photophysical properties make these nucleosides attractive candidates for enzymatic incorporation and biochemical assays. While 6-azauridine triphosphate is known to be poorly tolerated by polymerases in RNA synthesis, we illustrate that conjugating a thiophene ring at position 5 overcomes such limitations, facilitating its T7 RNA polymerase-mediated in vitro transcription incorporation into RNA constructs. We further show that the modified transcripts can be ligated to longer oligonucleotides to form singly modified RNAs, as illustrated for an A-site hairpin model RNA construct, which was employed to visualize aminoglycoside antibiotics binding.
Figures




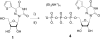
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

