Engineering orthogonal dual transcription factors for multi-input synthetic promoters
- PMID: 27982027
- PMCID: PMC5171851
- DOI: 10.1038/ncomms13858
Engineering orthogonal dual transcription factors for multi-input synthetic promoters
Abstract
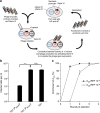
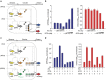
Synthetic biology has seen an explosive growth in the capability of engineering artificial gene circuits from transcription factors (TFs), particularly in bacteria. However, most artificial networks still employ the same core set of TFs (for example LacI, TetR and cI). The TFs mostly function via repression and it is difficult to integrate multiple inputs in promoter logic. Here we present to our knowledge the first set of dual activator-repressor switches for orthogonal logic gates, based on bacteriophage λ cI variants and multi-input promoter architectures. Our toolkit contains 12 TFs, flexibly operating as activators, repressors, dual activator-repressors or dual repressor-repressors, on up to 270 synthetic promoters. To engineer non cross-reacting cI variants, we design a new M13 phagemid-based system for the directed evolution of biomolecules. Because cI is used in so many synthetic biology projects, the new set of variants will easily slot into the existing projects of other groups, greatly expanding current engineering capacities.
Figures



References
-
- Elowitz M. B. & Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature 403, 335–338 (2000). - PubMed
-
- Guet C. C., Elowitz M. B., Hsing W. & Leibler S. Combinatorial synthesis of genetic networks. Science 296, 1466–1470 (2002). - PubMed
-
- Hasty J., Dolnik M., Rottschäfer V. & Collins J. J. Synthetic gene network for entraining and amplifying cellular oscillations. Phys. Rev. Lett. 88, 148101 (2002). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

