Insights into circular RNA biology
- PMID: 27982727
- PMCID: PMC5680708
- DOI: 10.1080/15476286.2016.1271524
Insights into circular RNA biology
Abstract
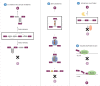
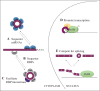
Circular RNAs (circRNAs) are a novel class of non-coding RNA characterized by a covalently closed-loop structure generated through a special type of alternative splicing termed backsplicing. CircRNAs are emerging as a heterogeneous class of molecules involved in modulating gene expression by regulation of transcription, protein and miRNA functions. CircRNA expression is cell type and tissue specific and can be largely independent of the expression level of the linear host gene, indicating that regulation of expression might be an important aspect with regard to control of circRNA function. In this review, a brief introduction to the characteristics that define a circRNA will be given followed by a discussion of putative biogenesis pathways and modulators of circRNA expression as well as of the stage at which circRNA formation takes place. A brief summary of circRNA functions will also be provided and lastly, an outlook with a focus on unanswered questions regarding circRNA biology will be included.
Keywords: Alternative splicing; backsplicing; biogenesis; circRNA; circular RNA; inverted repeats.
Figures

References
-
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al.. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013; 495:333-8; PMID:23446348; https://doi.org/10.1038/nature11928 - DOI - PubMed
-
- Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 2012; 7:e30733; PMID:22319583; https://doi.org/10.1371/journal.pone.0030733 - DOI - PMC - PubMed
-
- Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J 2011; 30:4414-22; PMID:21964070; https://doi.org/10.1038/emboj.2011.359 - DOI - PMC - PubMed
-
- Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol 2014; 15:409; PMID:25070500; https://doi.org/10.1186/s13059-014-0409-z - DOI - PMC - PubMed
-
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014; 32:453-61; PMID:24811520; https://doi.org/10.1038/nbt.2890 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
