Hepatitis E Virus in Wild Boars and Spillover Infection in Red and Roe Deer, Germany, 2013-2015
- PMID: 27983488
- PMCID: PMC5176221
- DOI: 10.3201/eid2301.161169
Hepatitis E Virus in Wild Boars and Spillover Infection in Red and Roe Deer, Germany, 2013-2015
Abstract
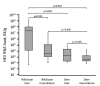
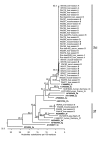
To determine animal hepatitis E virus (HEV) reservoirs, we analyzed serologic and molecular markers of HEV infection among wild animals in Germany. We detected HEV genotype 3 strains in inner organs and muscle tissues of a high percentage of wild boars and a lower percentage of deer, indicating a risk for foodborne infection of humans.
Keywords: Germany; Hepatitis E virus; fallow deer; red deer; roe deer; spillover; viruses; wild boar; zoonoses.
Figures
Similar articles
-
Prevalence of hepatitis E virus (HEV) Infection in wild boars and deer and genetic identification of a genotype 3 HEV from a boar in Japan.J Clin Microbiol. 2004 Nov;42(11):5371-4. doi: 10.1128/JCM.42.11.5371-5374.2004. J Clin Microbiol. 2004. PMID: 15528746 Free PMC article.
-
Prevalence and phylogenetic analysis of hepatitis E virus in pigs, wild boars, roe deer, red deer and moose in Lithuania.Acta Vet Scand. 2018 Feb 23;60(1):13. doi: 10.1186/s13028-018-0367-7. Acta Vet Scand. 2018. PMID: 29471843 Free PMC article.
-
Seroprevalence and molecular detection of hepatitis E virus in wild boar and red deer in The Netherlands.J Virol Methods. 2010 Sep;168(1-2):197-206. doi: 10.1016/j.jviromet.2010.05.014. Epub 2010 May 25. J Virol Methods. 2010. PMID: 20510298
-
The molecular epidemiology of hepatitis E virus infection.Virus Res. 2011 Oct;161(1):31-9. doi: 10.1016/j.virusres.2011.04.030. Epub 2011 May 11. Virus Res. 2011. PMID: 21600939 Review.
-
Epidemiology of hepatitis E in South-East Europe in the "One Health" concept.World J Gastroenterol. 2019 Jul 7;25(25):3168-3182. doi: 10.3748/wjg.v25.i25.3168. World J Gastroenterol. 2019. PMID: 31333309 Free PMC article. Review.
Cited by
-
Detection of Hepatitis E Virus in Livers and Muscle Tissues of Wild Boars in Italy.Food Environ Virol. 2020 Mar;12(1):1-8. doi: 10.1007/s12560-019-09405-0. Epub 2019 Sep 10. Food Environ Virol. 2020. PMID: 31506837
-
Molecular characterisation of a rabbit Hepatitis E Virus strain detected in a chronically HEV-infected individual from Germany.One Health. 2023 Mar 22;16:100528. doi: 10.1016/j.onehlt.2023.100528. eCollection 2023 Jun. One Health. 2023. PMID: 37363232 Free PMC article.
-
Increasing Hepatitis E Virus Seroprevalence in Domestic Pigs and Wild Boar in Bulgaria.Animals (Basel). 2020 Aug 28;10(9):1521. doi: 10.3390/ani10091521. Animals (Basel). 2020. PMID: 32872096 Free PMC article.
-
Standardised Sampling Approach for Investigating Pathogens or Environmental Chemicals in Wild Game at Community Hunts.Animals (Basel). 2022 Mar 31;12(7):888. doi: 10.3390/ani12070888. Animals (Basel). 2022. PMID: 35405877 Free PMC article.
-
Open reading frame 3 protein of hepatitis E virus: Multi-function protein with endless potential.World J Gastroenterol. 2021 May 28;27(20):2458-2473. doi: 10.3748/wjg.v27.i20.2458. World J Gastroenterol. 2021. PMID: 34092969 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

