A gene expression inflammatory signature specifically predicts multiple myeloma evolution and patients survival
- PMID: 27983725
- PMCID: PMC5223153
- DOI: 10.1038/bcj.2016.118
A gene expression inflammatory signature specifically predicts multiple myeloma evolution and patients survival
Abstract
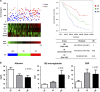
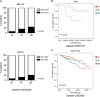
Multiple myeloma (MM) is closely dependent on cross-talk between malignant plasma cells and cellular components of the inflammatory/immunosuppressive bone marrow milieu, which promotes disease progression, drug resistance, neo-angiogenesis, bone destruction and immune-impairment. We investigated the relevance of inflammatory genes in predicting disease evolution and patient survival. A bioinformatics study by Ingenuity Pathway Analysis on gene expression profiling dataset of monoclonal gammopathy of undetermined significance, smoldering and symptomatic-MM, identified inflammatory and cytokine/chemokine pathways as the most progressively affected during disease evolution. We then selected 20 candidate genes involved in B-cell inflammation and we investigated their role in predicting clinical outcome, through univariate and multivariate analyses (log-rank test, logistic regression and Cox-regression model). We defined an 8-genes signature (IL8, IL10, IL17A, CCL3, CCL5, VEGFA, EBI3 and NOS2) identifying each condition (MGUS/smoldering/symptomatic-MM) with 84% accuracy. Moreover, six genes (IFNG, IL2, LTA, CCL2, VEGFA, CCL3) were found independently correlated with patients' survival. Patients whose MM cells expressed high levels of Th1 cytokines (IFNG/LTA/IL2/CCL2) and low levels of CCL3 and VEGFA, experienced the longest survival. On these six genes, we built a prognostic risk score that was validated in three additional independent datasets. In this study, we provide proof-of-concept that inflammation has a critical role in MM patient progression and survival. The inflammatory-gene prognostic signature validated in different datasets clearly indicates novel opportunities for personalized anti-MM treatment.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

