Internal Structure and Preferential Protein Binding of Colloidal Aggregates
- PMID: 27983786
- PMCID: PMC5491102
- DOI: 10.1021/acschembio.6b00791
Internal Structure and Preferential Protein Binding of Colloidal Aggregates
Abstract
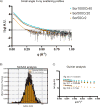
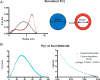
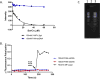
Colloidal aggregates of small molecules are the most common artifact in early drug discovery, sequestering and inhibiting target proteins without specificity. Understanding their structure and mechanism has been crucial to developing tools to control for, and occasionally even exploit, these particles. Unfortunately, their polydispersity and transient stability have prevented exploration of certain elementary properties, such as how they pack. Dye-stabilized colloidal aggregates exhibit enhanced homogeneity and stability when compared to conventional colloidal aggregates, enabling investigation of some of these properties. By small-angle X-ray scattering and multiangle light scattering, pair distance distribution functions suggest that the dye-stabilized colloids are filled, not hollow, spheres. Stability of the coformulated colloids enabled investigation of their preference for binding DNA, peptides, or folded proteins, and their ability to purify one from the other. The coformulated colloids showed little ability to bind DNA. Correspondingly, the colloids preferentially sequestered protein from even a 1600-fold excess of peptides that are themselves the result of a digest of the same protein. This may reflect the avidity advantage that a protein has in a surface-to-surface interaction with the colloids. For the first time, colloids could be shown to have preferences of up to 90-fold for particular proteins over others. Loaded onto the colloids, bound enzyme could be spun down, resuspended, and released back into buffer, regaining most of its activity. Implications of these observations for colloid mechanisms and utility will be considered.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
-
- McGovern SL, Caselli E, Grigorieff N, Shoichet BK. A common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening. J Med Chem. 2002;45:1712–1722. - PubMed
-
- McGovern SL, Helfand B, Feng B, Shoichet BK. A Specific Mechanism for Non-Specific Inhibition. J Med Chem. 2003;46:4265–4272. - PubMed
-
- Ryan AJ, Gray NM, Lowe PN, Chung C. Effect of Detergent on “Promiscuous” Inhibitors. J Med Chem. 2003;46:3448–3451. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

