CDK Substrate Phosphorylation and Ordering the Cell Cycle
- PMID: 27984725
- PMCID: PMC5161751
- DOI: 10.1016/j.cell.2016.11.034
CDK Substrate Phosphorylation and Ordering the Cell Cycle
Abstract
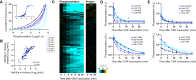
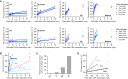
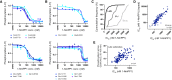
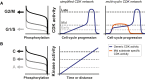
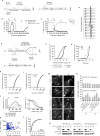
S phase and mitotic onset are brought about by the action of multiple different cyclin-CDK complexes. However, it has been suggested that changes in the total level of CDK kinase activity, rather than substrate specificity, drive the temporal ordering of S phase and mitosis. Here, we present a phosphoproteomics-based systems analysis of CDK substrates in fission yeast and demonstrate that the phosphorylation of different CDK substrates can be temporally ordered during the cell cycle by a single cyclin-CDK. This is achieved by rising CDK activity and the differential sensitivity of substrates to CDK activity over a wide dynamic range. This is combined with rapid phosphorylation turnover to generate clearly resolved substrate-specific activity thresholds, which in turn ensures the appropriate ordering of downstream cell-cycle events. Comparative analysis with wild-type cells expressing multiple cyclin-CDK complexes reveals how cyclin-substrate specificity works alongside activity thresholds to fine-tune the patterns of substrate phosphorylation.
Keywords: CDK; S phase; cell cycle; cyclin-dependent kinase; kinase; mitosis; phosphoproteomics; phosphorylation.
Copyright © 2016 The Francis Crick Institute. Published by Elsevier Inc. All rights reserved.
Figures










References
-
- Bähler J., Wu J.Q., Longtine M.S., Shah N.G., McKenzie A., 3rd, Steever A.B., Wach A., Philippsen P., Pringle J.R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. - PubMed
-
- Bishop A.C., Ubersax J.A., Petsch D.T., Matheos D.P., Gray N.S., Blethrow J., Shimizu E., Tsien J.Z., Schultz P.G., Rose M.D. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

