Development of a cancer-marker activated enzymatic switch from the herpes simplex virus thymidine kinase
- PMID: 27986921
- PMCID: PMC6080848
- DOI: 10.1093/protein/gzw067
Development of a cancer-marker activated enzymatic switch from the herpes simplex virus thymidine kinase
Abstract
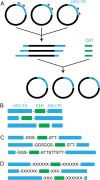
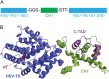
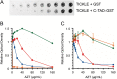
Discovery of new cancer biomarkers and advances in targeted gene delivery mechanisms have made gene-directed enzyme prodrug therapy (GDEPT) an attractive method for treating cancer. Recent focus has been placed on increasing target specificity of gene delivery systems and reducing toxicity in non-cancer cells in order to make GDEPT viable. To help address this challenge, we have developed an enzymatic switch that confers higher prodrug toxicity in the presence of a cancer marker. The enzymatic switch was derived from the herpes simplex virus thymidine kinase (HSV-TK) fused to the CH1 domain of the p300 protein. The CH1 domain binds to the C-terminal transactivation domain (C-TAD) of the cancer marker hypoxia inducible factor 1α. The switch was developed using a directed evolution approach that evaluated a large library of HSV-TK/CH1 fusions using a negative selection for azidothymidine (AZT) toxicity and a positive selection for dT phosphorylation. The identified switch, dubbed TICKLE (Trigger-Induced Cell-Killing Lethal-Enzyme), confers a 4-fold increase in AZT toxicity in the presence of C-TAD. The broad substrate specificity exhibited by HSV-TK makes TICKLE an appealing prospect for testing in medical imaging and cancer therapy, while establishing a foundation for further engineering of nucleoside kinase protein switches.
Keywords: HSV; directed evolution; herpes simplex virus thymidine kinase; protein switch; thymidine kinase.
© The Author 2016. Published by Oxford University Press. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

