Simultaneous Emergence of Multidrug-Resistant Candida auris on 3 Continents Confirmed by Whole-Genome Sequencing and Epidemiological Analyses
- PMID: 27988485
- PMCID: PMC5215215
- DOI: 10.1093/cid/ciw691
Simultaneous Emergence of Multidrug-Resistant Candida auris on 3 Continents Confirmed by Whole-Genome Sequencing and Epidemiological Analyses
Erratum in
-
Erratum.Clin Infect Dis. 2018 Aug 31;67(6):987. doi: 10.1093/cid/ciy333. Clin Infect Dis. 2018. PMID: 31206152 Free PMC article. No abstract available.
Abstract
Background: Candida auris, a multidrug-resistant yeast that causes invasive infections, was first described in 2009 in Japan and has since been reported from several countries.
Methods: To understand the global emergence and epidemiology of C. auris, we obtained isolates from 54 patients with C. auris infection from Pakistan, India, South Africa, and Venezuela during 2012-2015 and the type specimen from Japan. Patient information was available for 41 of the isolates. We conducted antifungal susceptibility testing and whole-genome sequencing (WGS).
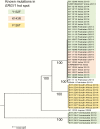
Results: Available clinical information revealed that 41% of patients had diabetes mellitus, 51% had undergone recent surgery, 73% had a central venous catheter, and 41% were receiving systemic antifungal therapy when C. auris was isolated. The median time from admission to infection was 19 days (interquartile range, 9-36 days), 61% of patients had bloodstream infection, and 59% died. Using stringent break points, 93% of isolates were resistant to fluconazole, 35% to amphotericin B, and 7% to echinocandins; 41% were resistant to 2 antifungal classes and 4% were resistant to 3 classes. WGS demonstrated that isolates were grouped into unique clades by geographic region. Clades were separated by thousands of single-nucleotide polymorphisms, but within each clade isolates were clonal. Different mutations in ERG11 were associated with azole resistance in each geographic clade.
Conclusions: C. auris is an emerging healthcare-associated pathogen associated with high mortality. Treatment options are limited, due to antifungal resistance. WGS analysis suggests nearly simultaneous, and recent, independent emergence of different clonal populations on 3 continents. Risk factors and transmission mechanisms need to be elucidated to guide control measures.
Keywords: Candida auris; amphotericin B resistance; candidemia; fluconazole resistance; whole genome sequence typing..
Published by Oxford University Press for the Infectious Diseases Society of America 2016. This work is written by (a) US Government employee(s) and is in the public domain in the US.
Figures
Comment in
-
Emergence of Candida auris: An International Call to Arms.Clin Infect Dis. 2017 Jan 15;64(2):141-143. doi: 10.1093/cid/ciw696. Epub 2016 Oct 20. Clin Infect Dis. 2017. PMID: 27989986 No abstract available.
References
-
- Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol 2009; 53:41–4. - PubMed
-
- Kim MN, Shin JH, Sung H, et al. Candida haemulonii and closely related species at 5 university hospitals in Korea: identification, antifungal susceptibility, and clinical features. Clin Infect Dis 2009; 48:e57–61. - PubMed
-
- Chakrabarti A, Sood P, Rudramurthy SM, et al. Incidence, characteristics and outcome of ICU-acquired candidemia in India. Intensive Care Med 2015; 41:285–95. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

