How the Eukaryotic Replisome Achieves Rapid and Efficient DNA Replication
- PMID: 27989442
- PMCID: PMC5222725
- DOI: 10.1016/j.molcel.2016.11.017
How the Eukaryotic Replisome Achieves Rapid and Efficient DNA Replication
Abstract
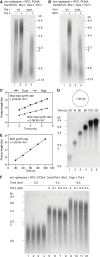
The eukaryotic replisome is a molecular machine that coordinates the Cdc45-MCM-GINS (CMG) replicative DNA helicase with DNA polymerases α, δ, and ε and other proteins to copy the leading- and lagging-strand templates at rates between 1 and 2 kb min-1. We have now reconstituted this sophisticated machine with purified proteins, beginning with regulated CMG assembly and activation. We show that replisome-associated factors Mrc1 and Csm3/Tof1 are crucial for in vivo rates of replisome progression. Additionally, maximal rates only occur when DNA polymerase ε catalyzes leading-strand synthesis together with its processivity factor PCNA. DNA polymerase δ can support leading-strand synthesis, but at slower rates. DNA polymerase δ is required for lagging-strand synthesis, but surprisingly also plays a role in establishing leading-strand synthesis, before DNA polymerase ε engagement. We propose that switching between these DNA polymerases also contributes to leading-strand synthesis under conditions of replicative stress.
Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
Figures







References
-
- Chilkova O., Stenlund P., Isoz I., Stith C.M., Grabowski P., Lundström E.B., Burgers P.M., Johansson E. The eukaryotic leading and lagging strand DNA polymerases are loaded onto primer-ends via separate mechanisms but have comparable processivity in the presence of PCNA. Nucleic Acids Res. 2007;35:6588–6597. - PMC - PubMed
-
- Clausen A.R., Lujan S.A., Burkholder A.B., Orebaugh C.D., Williams J.S., Clausen M.F., Malc E.P., Mieczkowski P.A., Fargo D.C., Smith D.J., Kunkel T.A. Tracking replication enzymology in vivo by genome-wide mapping of ribonucleotide incorporation. Nat. Struct. Mol. Biol. 2015;22:185–191. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

