Transcriptional architecture of the mammalian circadian clock
- PMID: 27990019
- PMCID: PMC5501165
- DOI: 10.1038/nrg.2016.150
Transcriptional architecture of the mammalian circadian clock
Abstract
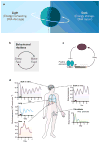
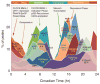
Circadian clocks are endogenous oscillators that control 24-hour physiological and behavioural processes in organisms. These cell-autonomous clocks are composed of a transcription-translation-based autoregulatory feedback loop. With the development of next-generation sequencing approaches, biochemical and genomic insights into circadian function have recently come into focus. Genome-wide analyses of the clock transcriptional feedback loop have revealed a global circadian regulation of processes such as transcription factor occupancy, RNA polymerase II recruitment and initiation, nascent transcription, and chromatin remodelling. The genomic targets of circadian clocks are pervasive and are intimately linked to the regulation of metabolism, cell growth and physiology.
Conflict of interest statement
The author declares competing interests. See the article online for details.
Figures





References
-
- Pittendrigh CS. Temporal organization: reflections of a Darwinian clock-watcher. Annu Rev Physiol. 1993;55:16–54. - PubMed
-
- Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. - PubMed
-
- Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. - PubMed
-
- Young MW, Kay SA. Time zones: a comparative genetics of circadian clocks. Nat Rev Genet. 2001;2:702–715. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

