Transcriptional Regulation of Tetrapyrrole Biosynthesis in Arabidopsis thaliana
- PMID: 27990150
- PMCID: PMC5130987
- DOI: 10.3389/fpls.2016.01811
Transcriptional Regulation of Tetrapyrrole Biosynthesis in Arabidopsis thaliana
Abstract
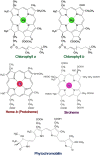
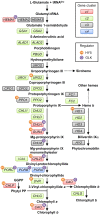
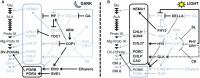
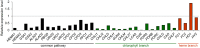
Biosynthesis of chlorophyll (Chl) involves many enzymatic reactions that share several first steps for biosynthesis of other tetrapyrroles such as heme, siroheme, and phycobilins. Chl allows photosynthetic organisms to capture light energy for photosynthesis but with simultaneous threat of photooxidative damage to cells. To prevent photodamage by Chl and its highly photoreactive intermediates, photosynthetic organisms have developed multiple levels of regulatory mechanisms to coordinate tetrapyrrole biosynthesis (TPB) with the formation of photosynthetic and photoprotective systems and to fine-tune the metabolic flow with the varying needs of Chl and other tetrapyrroles under various developmental and environmental conditions. Among a wide range of regulatory mechanisms of TPB, this review summarizes transcriptional regulation of TPB genes during plant development, with focusing on several transcription factors characterized in Arabidopsis thaliana. Key TPB genes are tightly coexpressed with other photosynthesis-associated nuclear genes and are induced by light, oscillate in a diurnal and circadian manner, are coordinated with developmental and nutritional status, and are strongly downregulated in response to arrested chloroplast biogenesis. LONG HYPOCOTYL 5 and PHYTOCHROME-INTERACTING FACTORs, which are positive and negative transcription factors with a wide range of light signaling, respectively, target many TPB genes for light and circadian regulation. GOLDEN2-LIKE transcription factors directly regulate key TPB genes to fine-tune the formation of the photosynthetic apparatus with chloroplast functionality. Some transcription factors such as FAR-RED ELONGATED HYPOCOTYL3, REVEILLE1, and scarecrow-like transcription factors may directly regulate some specific TPB genes, whereas other factors such as GATA transcription factors are likely to regulate TPB genes in an indirect manner. Comprehensive transcriptional analyses of TPB genes and detailed characterization of key transcriptional regulators help us obtain a whole picture of transcriptional control of TPB in response to environmental and endogenous cues.
Keywords: Arabidopsis thaliana; chlorophyll; chloroplast; gene expression; heme; photosynthesis; porphyrin; tetrapyrrole.
Figures

References
-
- Albus C. A., Salinas A., Czarnecki O., Kahlau S., Rothbart M., Thiele W., et al. (2012). LCAA, a novel factor required for magnesium protoporphyrin monomethylester cyclase accumulation and feedback control of aminolevulinic acid biosynthesis in tobacco. Plant Physiol. 160 1923–1939. 10.1104/pp.112.206045 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

