The Identification and Characterization of Two Novel Epitopes on the Nucleocapsid Protein of the Porcine Epidemic Diarrhea Virus
- PMID: 27991537
- PMCID: PMC5171872
- DOI: 10.1038/srep39010
The Identification and Characterization of Two Novel Epitopes on the Nucleocapsid Protein of the Porcine Epidemic Diarrhea Virus
Abstract
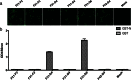
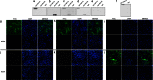
Porcine epidemic diarrhea virus (PEDV) is a highly contagious coronavirus that causes severe diarrhea and death, particularly in neonatal piglets. The nucleocapsid protein (N protein) of PEDV presents strong immunogenicity and contributes to the cross-reactivity between PEDV and TGEV. However, the characterization of epitopes on the PEDV N protein remains largely unknown. Here, two monoclonal antibodies (MAbs) specific to the N protein of a PEDV strain, FJzz1/2011, were generated and screened against a partially overlapping library of 24 GST-fusion N protein-truncated constructs. We confirmed that residues 18-133 (designated NEP-D4) and residues 252-262 (designated NEP-D6) were the epitopes targeted by MAbs PN-D4 and PN-D6, respectively. Sequence analysis revealed that these two epitopes were highly conserved among PEDV strains but were significantly different from other members of the Coronavirinae subfamily. Western blot analysis showed that they could be specifically recognized by PEDV antisera but could not be recognized by TGEV hyperimmune antisera. Indirect immunofluorescence (IFA) assays confirmed no cross-reaction between these two MAbs and TGEV. In addition, the freeze-thaw cycle and protease treatment results indicated that NEP-D4 was intrinsically disordered. All these results suggest that these two novel epitopes and their cognate MAbs could serve as the basis for the development of precise diagnostic assays for PEDV.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
-
- Duarte M., Gelfi J., Lambert P., Rasschaert D. & Laude H. Genome organization of porcine epidemic diarrhoea virus. Adv. Exp. Med. Biol. 342, 55–60 (1993). - PubMed
-
- Enjuanes L., Siddell S. & Spaan W. In Coronaviruses and Arteriviruses (eds Bridgen A., Kocherhans R., Tobler K., Carvajal A. & Ackermann M.) 781–786 (Springer, 1998).
-
- Kusanagi K. et al. Isolation and serial propagation of porcine epidemic diarrhea virus in cell cultures and partial characterization of the isolate. J. Vet. Med. Sci. 54, 313–318 (1992). - PubMed
-
- Wood E. N. An apparently new syndrome of porcine epidemic diarrhoea. Vet. Rec. 100, 243–244 (1977). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

