Linking genomic reorganization to tumor initiation via the giant cell cycle
- PMID: 27991913
- PMCID: PMC5177773
- DOI: 10.1038/oncsis.2016.75
Linking genomic reorganization to tumor initiation via the giant cell cycle
Abstract
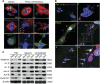
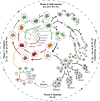
To investigate the mechanisms underlying our recent paradoxical finding that mitotically incapacitated and genomically unstable polyploid giant cancer cells (PGCCs) are capable of tumor initiation, we labeled ovarian cancer cells with α-tubulin fused to green fluorescent protein, histone-2B fused to red fluorescent protein and FUCCI (fluorescent ubiquitination cell cycle indicator), and tracked the spatial and time-dependent change in spindle and chromosomal dynamics of PGCCs using live-cell fluorescence time-lapse recording. We found that single-dose (500 nm) treatment with paclitaxel paradoxically initiated endoreplication to form PGCCs after massive cell death. The resulting PGCCs continued self-renewal via endoreplication and further divided by nuclear budding or fragmentation; the small daughter nuclei then acquired cytoplasm, split off from the giant mother cells and acquired competency in mitosis. FUCCI showed that PGCCs divided via truncated endoreplication cell cycle (endocycle or endomitosis). Confocal microscopy showed that PGCCs had pronounced nuclear fragmentation and lacked expression of key mitotic proteins. PGCC-derived daughter cells were capable of long-term proliferation and acquired numerous new genome/chromosome alterations demonstrated by spectral karyotyping. These data prompt us to conceptualize a giant cell cycle composed of four distinct but overlapping phases, initiation, self-renewal, termination and stability. The giant cell cycle may represent a fundamental cellular mechanism to initiate genomic reorganization to generate new tumor-initiating cells in response to chemotherapy-induced stress and contributes to disease relapse.
Figures




References
-
- Edgar BA, Zielke N, Gutierrez C. Endocycles: a recurrent evolutionary innovation for post-mitotic cell growth. Nat Rev Mol Cell Biol 2014; 15: 197–210. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

