The histone demethylase UTX regulates the lineage-specific epigenetic program of invariant natural killer T cells
- PMID: 27992400
- PMCID: PMC5247321
- DOI: 10.1038/ni.3644
The histone demethylase UTX regulates the lineage-specific epigenetic program of invariant natural killer T cells
Abstract
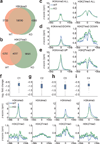
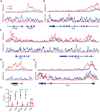
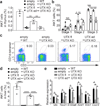
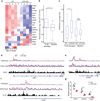
Invariant natural killer T cells (iNKT cells) are innate-like lymphocytes that protect against infection, autoimmune disease and cancer. However, little is known about the epigenetic regulation of iNKT cell development. Here we found that the H3K27me3 histone demethylase UTX was an essential cell-intrinsic factor that controlled an iNKT-cell lineage-specific gene-expression program and epigenetic landscape in a demethylase-activity-dependent manner. UTX-deficient iNKT cells exhibited impaired expression of iNKT cell signature genes due to a decrease in activation-associated H3K4me3 marks and an increase in repressive H3K27me3 marks within the promoters occupied by UTX. We found that JunB regulated iNKT cell development and that the expression of genes that were targets of both JunB and the iNKT cell master transcription factor PLZF was UTX dependent. We identified iNKT cell super-enhancers and demonstrated that UTX-mediated regulation of super-enhancer accessibility was a key mechanism for commitment to the iNKT cell lineage. Our findings reveal how UTX regulates the development of iNKT cells through multiple epigenetic mechanisms.
Figures



Comment in
-
iNKT cells need UTX-tra demethylation.Nat Immunol. 2017 Jan 19;18(2):148-150. doi: 10.1038/ni.3663. Nat Immunol. 2017. PMID: 28102220 No abstract available.
References
-
- Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu. Rev. Immunol. 2007;25:297–336. - PubMed
-
- Kawano T, et al. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science. 1997;278:1626–1629. - PubMed
-
- Cui J, et al. Requirement for Valpha14 NKT cells in IL-12-mediated rejection of tumors. Science. 1997;278:1623–1626. - PubMed
References for Methods
-
- Passegue E, Wagner EF, Weissman IL. JunB deficiency leads to a myeloproliferative disorder arising from hematopoietic stem cells. Cell. 2004;119:431–443. - PubMed
-
- Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. - PubMed
-
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. - PubMed
-
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 Article3. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

