A genome-wide CRISPR screen identifies a restricted set of HIV host dependency factors
- PMID: 27992415
- PMCID: PMC5511375
- DOI: 10.1038/ng.3741
A genome-wide CRISPR screen identifies a restricted set of HIV host dependency factors
Abstract
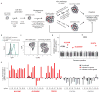
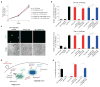
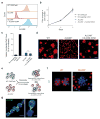
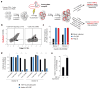
Host proteins are essential for HIV entry and replication and can be important nonviral therapeutic targets. Large-scale RNA interference (RNAi)-based screens have identified nearly a thousand candidate host factors, but there is little agreement among studies and few factors have been validated. Here we demonstrate that a genome-wide CRISPR-based screen identifies host factors in a physiologically relevant cell system. We identify five factors, including the HIV co-receptors CD4 and CCR5, that are required for HIV infection yet are dispensable for cellular proliferation and viability. Tyrosylprotein sulfotransferase 2 (TPST2) and solute carrier family 35 member B2 (SLC35B2) function in a common pathway to sulfate CCR5 on extracellular tyrosine residues, facilitating CCR5 recognition by the HIV envelope. Activated leukocyte cell adhesion molecule (ALCAM) mediates cell aggregation, which is required for cell-to-cell HIV transmission. We validated these pathways in primary human CD4+ T cells through Cas9-mediated knockout and antibody blockade. Our findings indicate that HIV infection and replication rely on a limited set of host-dispensable genes and suggest that these pathways can be studied for therapeutic intervention.
Conflict of interest statement
T.W., D.M.S., and E.S.L. are inventors on a patent application for functional genomics using the CRISPR-Cas system (US 15/141,348), T.W and D.M.S are founders of KSQ Therapeutics, a CRISPR functional genomics company, and D.M.S is a scientific advisor for KSQ Therapeutics. A patent has been filed on the use of Cas9 RNPs to edit the genome of human primary T cells (A.M. and K.S.). A.M. serves as an advisor to Juno Therapeutics and the Marson lab has had sponsored research agreements with Juno Therapeutics and Epinomics.
Figures


Comment in
-
Picking the Survivor! CRISPR Reveals HIV Dependency Factors.Trends Microbiol. 2017 Apr;25(4):243-245. doi: 10.1016/j.tim.2017.02.004. Epub 2017 Feb 21. Trends Microbiol. 2017. PMID: 28233621
References
-
- Friedrich BM, Dziuba N, Li G, Endsley MA, et al. Host factors mediating HIV-1 replication. Virus Res. 2011;161:101–114. - PubMed
-
- Fätkenheuer G, Pozniak AL, Johnson MA, Plettenberg A, et al. Efficacy of short-term monotherapy with maraviroc, a new CCR5 antagonist, in patients infected with HIV-1. Nat Med. 2005;11:1170–1172. - PubMed
-
- Hütter G, Nowak D, Mossner M, Ganepola S, et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med. 2009;360:692–698. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

