Between the Balkans and the Baltic: Phylogeography of a Common Vole Mitochondrial DNA Lineage Limited to Central Europe
- PMID: 27992546
- PMCID: PMC5161492
- DOI: 10.1371/journal.pone.0168621
Between the Balkans and the Baltic: Phylogeography of a Common Vole Mitochondrial DNA Lineage Limited to Central Europe
Abstract
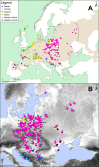
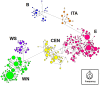
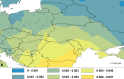
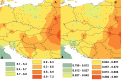
The common vole (Microtus arvalis) has been a model species of small mammal for studying end-glacial colonization history. In the present study we expanded the sampling from central and eastern Europe, analyzing contemporary genetic structure to identify the role of a potential 'northern glacial refugium', i.e. a refugium at a higher latitude than the traditional Mediterranean refugia. Altogether we analyzed 786 cytochrome b (cytb) sequences (representing mitochondrial DNA; mtDNA) from the whole of Europe, adding 177 new sequences from central and eastern Europe, and we conducted analyses on eight microsatellite loci for 499 individuals (representing nuclear DNA) from central and eastern Europe, adding data on 311 new specimens. Our new data fill gaps in the vicinity of the Carpathian Mountains, the potential northern refugium, such that there is now dense sampling from the Balkans to the Baltic Sea. Here we present evidence that the Eastern mtDNA lineage of the common vole was present in the vicinity of this Carpathian refugium during the Last Glacial Maximum and the Younger Dryas. The Eastern lineage expanded from this refugium to the Baltic and shows low cytb nucleotide diversity in those most northerly parts of the distribution. Analyses of microsatellites revealed a similar pattern but also showed little differentiation between all of the populations sampled in central and eastern Europe.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Avise JC. Phylogeography: The History and Formation of Species Cambridge (MA): Harvard University Press, 2000.
-
- Webb T, Bartlein PJ. Global changes during the last 3 million years: climatic controls and biotic responses. Annu Rev Ecol Evol Syst. 1992; 23: 141–173.
-
- Wysota W, Lankauf KR, Szmańda J, Chruścińska A, Oczkowski HL, Przegiętka KR. Chronology of the Vistulian (Weichselian) glacial events in the lower Vistula region, middle-north Poland. Geochronometria. 2002; 21: 137–142.
-
- Wysota W, Molewski P, Sokołowski RJ. Record of the Vistula ice lobe advances in the Late Weichselian glacial sequence in north-central Poland. Quat Int. 2009; 207: 26–41.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

