In situ structures of the genome and genome-delivery apparatus in a single-stranded RNA virus
- PMID: 27992877
- PMCID: PMC5701785
- DOI: 10.1038/nature20589
In situ structures of the genome and genome-delivery apparatus in a single-stranded RNA virus
Abstract
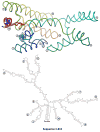
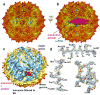
Packaging of the genome into a protein capsid and its subsequent delivery into a host cell are two fundamental processes in the life cycle of a virus. Unlike double-stranded DNA viruses, which pump their genome into a preformed capsid, single-stranded RNA (ssRNA) viruses, such as bacteriophage MS2, co-assemble their capsid with the genome; however, the structural basis of this co-assembly is poorly understood. MS2 infects Escherichia coli via the host 'sex pilus' (F-pilus); it was the first fully sequenced organism and is a model system for studies of translational gene regulation, RNA-protein interactions, and RNA virus assembly. Its positive-sense ssRNA genome of 3,569 bases is enclosed in a capsid with one maturation protein monomer and 89 coat protein dimers arranged in a T = 3 icosahedral lattice. The maturation protein is responsible for attaching the virus to an F-pilus and delivering the viral genome into the host during infection, but how the genome is organized and delivered is not known. Here we describe the MS2 structure at 3.6 Å resolution, determined by electron-counting cryo-electron microscopy (cryoEM) and asymmetric reconstruction. We traced approximately 80% of the backbone of the viral genome, built atomic models for 16 RNA stem-loops, and identified three conserved motifs of RNA-coat protein interactions among 15 of these stem-loops with diverse sequences. The stem-loop at the 3' end of the genome interacts extensively with the maturation protein, which, with just a six-helix bundle and a six-stranded β-sheet, forms a genome-delivery apparatus and joins 89 coat protein dimers to form a capsid. This atomic description of genome-capsid interactions in a spherical ssRNA virus provides insight into genome delivery via the host sex pilus and mechanisms underlying ssRNA-capsid co-assembly, and inspires speculation about the links between nucleoprotein complexes and the origins of viruses.
Conflict of interest statement
The authors declare no competing financial interests.
Figures











References
-
- Lander GC, et al. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006;312:1791–1795. - PubMed
-
- Catalano CE. Viral Genome Packaging Machines: Genetics, Structure, and Mechanism. Springer; US: 2005.
References in Methods
-
- Suloway C, et al. Automated molecular microscopy: the new Leginon system. Journal of structural biology. 2005;151:41–60. - PubMed
-
- Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. Journal of structural biology. 2003;142:334–347. - PubMed
-
- Kivioja T, Ravantti J, Verkhovsky A, Ukkonen E, Bamford D. Local average intensity-based method for identifying spherical particles in electron micrographs. Journal of structural biology. 2000;131:126–134. - PubMed
-
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. Journal of structural biology. 1999;128:82–97. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- DE023591/NH/NIH HHS/United States
- 1U24GM116792/NH/NIH HHS/United States
- R01 AI094386/AI/NIAID NIH HHS/United States
- CA177322/NH/NIH HHS/United States
- S10 OD018111/OD/NIH HHS/United States
- U24 GM116792/GM/NIGMS NIH HHS/United States
- R01 GM071940/GM/NIGMS NIH HHS/United States
- R01 DE023591/DE/NIDCR NIH HHS/United States
- DE025567/NH/NIH HHS/United States
- R01 DE025567/DE/NIDCR NIH HHS/United States
- GM071940/NH/NIH HHS/United States
- S10 RR023057/RR/NCRR NIH HHS/United States
- 1S10OD018111/NH/NIH HHS/United States
- AI094386/NH/NIH HHS/United States
- P01 CA177322/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

