Rational Design of Small Molecules Targeting Oncogenic Noncoding RNAs from Sequence
- PMID: 27993012
- PMCID: PMC5286924
- DOI: 10.1021/acs.accounts.6b00326
Rational Design of Small Molecules Targeting Oncogenic Noncoding RNAs from Sequence
Abstract
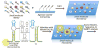
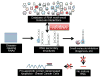
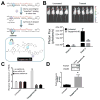
The discovery of RNA catalysis in the 1980s and the dissemination of the human genome sequence at the start of this century inspired investigations of the regulatory roles of noncoding RNAs in biology. In fact, the Encyclopedia of DNA Elements (ENCODE) project has shown that only 1-2% of the human genome encodes protein, yet 75% is transcribed into RNA. Functional studies both preceding and following the ENCODE project have shown that these noncoding RNAs have important roles in regulating gene expression, developmental timing, and other critical functions. RNA's diverse roles are often a consequence of the various folds that it adopts. The single-stranded nature of the biopolymer enables it to adopt intramolecular folds with noncanonical pairings to lower its free energy. These folds can be scaffolds to bind proteins or to form frameworks to interact with other RNAs. Not surprisingly, dysregulation of certain noncoding RNAs has been shown to be causative of disease. Given this as the background, it is easy to see why it would be useful to develop methods that target RNA and manipulate its biology in rational and predictable ways. The antisense approach has afforded strategies to target RNAs via Watson-Crick base pairing and has typically focused on targeting partially unstructured regions of RNA. Small molecule strategies to target RNA would be desirable not only because compounds could be lead optimized via medicinal chemistry but also because structured regions within an RNA of interest could be targeted to directly interfere with RNA folds that contribute to disease. Additionally, small molecules have historically been the most successful drug candidates. Until recently, the ability to design small molecules that target non-ribosomal RNAs has been elusive, creating the perception that they are "undruggable". In this Account, approaches to demystify targeting RNA with small molecules are described. Rather than bulk screening for compounds that bind to singular targets, which is the purview of the pharmaceutical industry and academic institutions with high throughput screening facilities, we focus on methods that allow for the rational design of small molecules toward biological RNAs. One enabling and foundational technology that has been developed is two-dimensional combinatorial screening (2DCS), a library-versus-library selection approach that allows the identification of the RNA motif binding preferences of small molecules from millions of combinations. A landscape map of the 2DCS-defined and annotated RNA motif-small molecule interactions is then placed into Inforna, a computational tool that allows one to mine these interactions against an RNA of interest or an entire transcriptome. Indeed, this approach has been enabled by tools to annotate RNA structure from sequence, an invaluable asset to the RNA community and this work, and has allowed for the rational identification of "druggable" RNAs in a target agnostic fashion.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
-
- Arrowsmith CH, Audia JE, Austin C, Baell J, Bennett J, Blagg J, Bountra C, Brennan PE, Brown PJ, Bunnage ME, Buser-Doepner C, Campbell RM, Carter AJ, Cohen P, Copeland RA, Cravatt B, Dahlin JL, Dhanak D, Edwards AM, Frederiksen M, Frye SV, Gray N, Grimshaw CE, Hepworth D, Howe T, Huber KV, Jin J, Knapp S, Kotz JD, Kruger RG, Lowe D, Mader MM, Marsden B, Mueller-Fahrnow A, Muller S, O’Hagan RC, Overington JP, Owen DR, Rosenberg SH, Roth B, Ross R, Schapira M, Schreiber SL, Shoichet B, Sundstrom M, Superti-Furga G, Taunton J, Toledo-Sherman L, Walpole C, Walters MA, Willson TM, Workman P, Young RN, Zuercher WJ. The promise and peril of chemical probes. Nat Chem Biol. 2015;11:536–541. doi: 10.1038/nchembio.1867. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

