Performance of Celera RUO integrase resistance assay across multiple HIV-1 subtypes
- PMID: 27993614
- PMCID: PMC5292360
- DOI: 10.1016/j.jviromet.2016.12.008
Performance of Celera RUO integrase resistance assay across multiple HIV-1 subtypes
Abstract
Background: HIV-1 sequence variation is a major obstacle to developing molecular based assays for multiple subtypes. This study sought to independently assess performance characteristics of the ViroSeq™ HIV-1 Integrase RUO Genotyping Kit (Celera, US) for samples of multiple different HIV-1 subtypes.
Methods: 264 samples were tested in the validation, 106 from integrase inhibitor naïve patients' sent for routine HIV-1 drug resistance testing after failing a 1st- or 2nd-line regimen, and 158 samples from an external virology quality assurance program (VQA). For the latter, 53 unique VQA samples were tested in two to five different laboratories to assess assay reproducibility. For all assays, viral RNA was extracted using the ViroSeq extraction module, reverse transcribed, and amplified in a one-step reaction. Four sequencing primers were used to span codons 1-288 of integrase. The Rega subtyping tool was used for subtype assignment. Integrase polymorphisms and mutations were determined as differences from the HXB2 sequence and by the Stanford database, respectively. Sequences obtained from the different laboratories were aligned and sequence homology determined.
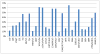
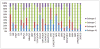
Results: HIV-1 RNA in the 264 samples ranged from 3.15 to 6.74logcopies/ml. Successful amplification was obtained for 97% of samples (n=256). The 8 samples that failed to amplify were subtype D (n=3), subtype C (n=1), CRF01_AE (n=1), subtype A1 (n=2), and an unassigned subtype (n=1). Of the 256 that successfully amplified samples, 203 (79%) were successfully sequenced with bidirectional coverage. Of the 53 unsuccessful samples, 13 (5%) failed sequencing and 40 (16%) did not have full bidirectional sequence, as a result of failure of sequencing primers: Primer A (n=1); Primer B (n=18); Primer C (n=1); Primer D (n=7) or short sequences (n=16). For the 135 VQA samples (30 unique samples) that were assayed by different laboratories, homology of the sequences obtained ranged from 92.1% to 100%. However, Laboratory 2 detected more mixtures (74%) compared to the other four laboratories, whereas Laboratory 1 detected the least number of mixtures (35%), likely due to differences between the labs in the methods of sequence analysis. Mutations associated with integrase resistance were observed in seven of the 106 (7%) clinical samples [one sample: Q148K; E138K; G140A; two samples: T97A and four samples: L74I]. Of the four samples with L74I, 3 were subtype G.
Conclusion: Of the total 264 samples tested, 243 (92%) of samples were able to be amplified and sequenced to generate an integrase genotype. Sequencing results were similar between the testing laboratories with the exception of mixture detection. Mutations associated with integrase inhibitor resistance were observed in only 7% of integrase inhibitor naive samples, and some of these mutations are likely to be due to subtype-specific polymorphisms rather than selection by an integrase inhibitor.
Keywords: HIV-1 drug resistance; Integrase; Non-subtype B.
Copyright © 2016 Elsevier B.V. All rights reserved.
Figures

Similar articles
-
Performance comparison of an in-house integrase genotyping assay versus the ViroSeq™ Integra48, and study of HIV-1 integrase polymorphisms in Hong Kong.J Clin Virol. 2013 Sep;58(1):299-302. doi: 10.1016/j.jcv.2013.06.040. Epub 2013 Jul 22. J Clin Virol. 2013. PMID: 23886504
-
Cross-clade simultaneous HIV drug resistance genotyping for reverse transcriptase, protease, and integrase inhibitor mutations by Illumina MiSeq.Retrovirology. 2014 Dec 23;11:122. doi: 10.1186/s12977-014-0122-8. Retrovirology. 2014. PMID: 25533166 Free PMC article.
-
Comparison of an in-house 'home-brew' and commercial ViroSeq integrase genotyping assays on HIV-1 subtype C samples.PLoS One. 2019 Nov 21;14(11):e0224292. doi: 10.1371/journal.pone.0224292. eCollection 2019. PLoS One. 2019. PMID: 31751353 Free PMC article.
-
Characterization and structural analysis of HIV-1 integrase conservation.AIDS Rev. 2009 Jan-Mar;11(1):17-29. AIDS Rev. 2009. PMID: 19290031 Review.
-
Resistance to HIV Integrase Inhibitors: About R263K and E157Q Mutations.Viruses. 2018 Jan 18;10(1):41. doi: 10.3390/v10010041. Viruses. 2018. PMID: 29346270 Free PMC article. Review.
Cited by
-
A Partially Multiplexed HIV Drug Resistance (HIVDR) Assay for Monitoring HIVDR Mutations of the Protease, Reverse-Transcriptase (PRRT), and Integrase (INT).Microbiol Spectr. 2022 Jun 29;10(3):e0177621. doi: 10.1128/spectrum.01776-21. Epub 2022 May 5. Microbiol Spectr. 2022. PMID: 35510849 Free PMC article.
-
Point-of-Care Tenofovir Urine Testing for the Prediction of Treatment Failure and Drug Resistance During Initial Treatment for Human Immunodeficiency Virus Type 1 (HIV-1) Infection.Clin Infect Dis. 2023 Feb 8;76(3):e553-e560. doi: 10.1093/cid/ciac755. Clin Infect Dis. 2023. PMID: 36136811 Free PMC article. Clinical Trial.
References
-
- Boulle A, Orrel C, Kaplan R, Van Cutsem G, McNally M, Hilderbrand K, Myer L, Egger M, Coetzee D, Maartens G, Wood R. Substitutions due to antiretroviral toxicity or contraindication in the first 3 years of antiretroviral therapy in a large South African cohort. Antiviral therapy. 2007;12:753–60. - PubMed
-
- Cahn P, Sued O. Raltegravir: a new antiretroviral class for salvage therapy. Lancet. 2007;369:1235–6. - PubMed
-
- Carr A, Cooper DA. Adverse effects of antiretroviral therapy. Lancet. 2000;356:1423–30. - PubMed
-
- Colquitt AR, Pham PA. The Hopkins HIV report: a bimonthly newsletter for healthcare providers. Vol. 19. Johns Hopkins University AIDS Service; 2007. Expanded access drug profile: raltegravir (RAL, MK-0518) pp. 11–2. - PubMed
-
- Grinsztejn B, Nguyen BY, Katlama C, Gatell JM, Lazzarin A, Vittecoq D, Gonzalez CJ, Chen J, Harvey CM, Isaacs RD, Protocol T. Safety and efficacy of the HIV-1 integrase inhibitor raltegravir (MK-0518) in treatment-experienced patients with multidrug-resistant virus: a phase II randomised controlled trial. Lancet. 2007;369:1261–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

