Human Apurinic/Apyrimidinic Endonuclease (APE1) Is Acetylated at DNA Damage Sites in Chromatin, and Acetylation Modulates Its DNA Repair Activity
- PMID: 27994014
- PMCID: PMC5335514
- DOI: 10.1128/MCB.00401-16
Human Apurinic/Apyrimidinic Endonuclease (APE1) Is Acetylated at DNA Damage Sites in Chromatin, and Acetylation Modulates Its DNA Repair Activity
Abstract
Apurinic/apyrimidinic (AP) sites, the most frequently formed DNA lesions in the genome, inhibit transcription and block replication. The primary enzyme that repairs AP sites in mammalian cells is the AP endonuclease (APE1), which functions through the base excision repair (BER) pathway. Although the mechanism by which APE1 repairs AP sites in vitro has been extensively investigated, it is largely unknown how APE1 repairs AP sites in cells. Here, we show that APE1 is acetylated (AcAPE1) after binding to the AP sites in chromatin and that AcAPE1 is exclusively present on chromatin throughout the cell cycle. Positive charges of acetylable lysine residues in the N-terminal domain of APE1 are essential for chromatin association. Acetylation-mediated neutralization of the positive charges of the lysine residues in the N-terminal domain of APE1 induces a conformational change; this in turn enhances the AP endonuclease activity of APE1. In the absence of APE1 acetylation, cells accumulated AP sites in the genome and showed higher sensitivity to DNA-damaging agents. Thus, mammalian cells, unlike Saccharomyces cerevisiae or Escherichia coli cells, require acetylation of APE1 for the efficient repair of AP sites and base damage in the genome. Our study reveals that APE1 acetylation is an integral part of the BER pathway for maintaining genomic integrity.
Keywords: AP site; APE1; DNA damage; acetylation; base excision repair; endogenous DNA damage.
Copyright © 2017 Roychoudhury et al.
Figures
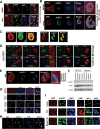

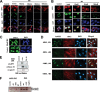

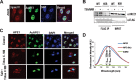
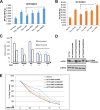

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
