Legionella pneumophila OxyR Is a Redundant Transcriptional Regulator That Contributes to Expression Control of the Two-Component CpxRA System
- PMID: 27994017
- PMCID: PMC5309917
- DOI: 10.1128/JB.00690-16
Legionella pneumophila OxyR Is a Redundant Transcriptional Regulator That Contributes to Expression Control of the Two-Component CpxRA System
Abstract
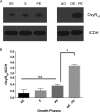
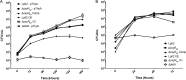
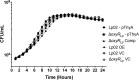
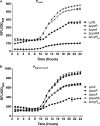
Nominally an environmental organism, Legionella pneumophila is an intracellular parasite of protozoa but is also the causative agent of the pneumonia termed Legionnaires' disease, which results from inhalation of aerosolized bacteria by susceptible humans. Coordination of gene expression by a number of identified regulatory factors, including OxyR, assists L. pneumophila in adapting to the stresses of changing environments. L. pneumophila OxyR (OxyRLp) is an ortholog of Escherichia coli OxyR; however, OxyRLp was shown elsewhere to be functionally divergent, such that it acts as a transcription regulator independently of the oxidative stress response. In this study, the use of improved gene deletion methods has enabled us to generate an unmarked in-frame deletion of oxyR in L. pneumophila Lack of OxyRLp did not affect in vitro growth or intracellular growth in Acanthamoeba castellanii protozoa and U937-derived macrophages. The expression of OxyRLp does not appear to be regulated by CpxR, even though purified recombinant CpxR bound a DNA sequence similar to that reported for CpxR elsewhere. Surprisingly, a lack of OxyRLp resulted in elevated activity of the promoters located upstream of icmR and the lpg1441-cpxA operon, and OxyRLp directly bound to these promoter regions, suggesting that OxyRLp is a direct repressor. Interestingly, a strain overexpressing OxyRLp demonstrated reduced intracellular growth in A. castellanii but not in U937-derived macrophages, suggesting that balanced expression control of the two-component CpxRA system is necessary for survival in protozoa. Taken together, this study suggests that OxyRLp is a functionally redundant transcriptional regulator in L. pneumophila under the conditions evaluated herein.IMPORTANCELegionella pneumophila is an environmental pathogen, with its transmission to the human host dependent upon its ability to replicate in protozoa and survive within its aquatic niche. Understanding the genetic factors that contribute to L. pneumophila survival within each of these unique environments will be key to limiting future point-source outbreaks of Legionnaires' disease. The transcriptional regulator L. pneumophila OxyR (OxyRLp) has been previously identified as a potential regulator of virulence traits warranting further investigation. This study demonstrated that oxyR is nonessential for L. pneumophila survival in vitro and in vivo via mutational analysis. While the mechanisms of how OxyRLp expression is regulated remain elusive, this study shows that OxyRLp negatively regulates the expression of the cpxRA two-component system necessary for intracellular survival in protozoa.
Keywords: CpxRA; Legionella pneumophila; OxyR; gene expression; intracellular pathogen; microbial genetics; protozoa; transcriptional regulation.
Copyright © 2017 American Society for Microbiology.
Figures




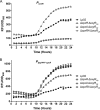
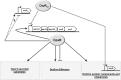
References
-
- Martinelli F, Carasi S, Scarcella C, Speziani F. 2001. Detection of Legionella pneumophila at thermal spas. New Microbiol 24:259–264. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

