Applicability of a computational design approach for synthetic riboswitches
- PMID: 27994029
- PMCID: PMC5397205
- DOI: 10.1093/nar/gkw1267
Applicability of a computational design approach for synthetic riboswitches
Abstract
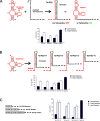
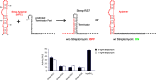
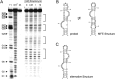
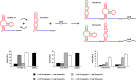
Riboswitches have gained attention as tools for synthetic biology, since they enable researchers to reprogram cells to sense and respond to exogenous molecules. In vitro evolutionary approaches produced numerous RNA aptamers that bind such small ligands, but their conversion into functional riboswitches remains difficult. We previously developed a computational approach for the design of synthetic theophylline riboswitches based on secondary structure prediction. These riboswitches have been constructed to regulate ligand-dependent transcription termination in Escherichia coli. Here, we test the usability of this design strategy by applying the approach to tetracycline and streptomycin aptamers. The resulting tetracycline riboswitches exhibit robust regulatory properties in vivo. Tandem fusions of these riboswitches with theophylline riboswitches represent logic gates responding to two different input signals. In contrast, the conversion of the streptomycin aptamer into functional riboswitches appears to be difficult. Investigations of the underlying aptamer secondary structure revealed differences between in silico prediction and structure probing. We conclude that only aptamers adopting the minimal free energy (MFE) structure are suitable targets for construction of synthetic riboswitches with design approaches based on equilibrium thermodynamics of RNA structures. Further improvements in the design strategy are required to implement aptamer structures not corresponding to the calculated MFE state.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Winkler W.C., Nahvi A., Breaker R.R.. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002; 419:952–956. - PubMed
-
- Nahvi A., Sudarsan N., Ebert M.S., Zou X., Brown K.L., Breaker R.R.. Genetic control by a metabolite binding mRNA. Chem. Biol. 2002; 9:1043. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

