Destabilization of the PCNA trimer mediated by its interaction with the NEIL1 DNA glycosylase
- PMID: 27994037
- PMCID: PMC5389659
- DOI: 10.1093/nar/gkw1282
Destabilization of the PCNA trimer mediated by its interaction with the NEIL1 DNA glycosylase
Abstract
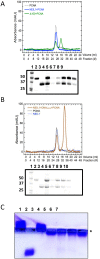
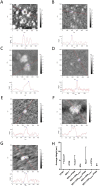
The base excision repair (BER) pathway repairs oxidized lesions in the DNA that result from reactive oxygen species generated in cells. If left unrepaired, these damaged DNA bases can disrupt cellular processes such as replication. NEIL1 is one of the 11 human DNA glycosylases that catalyze the first step of the BER pathway, i.e. recognition and excision of DNA lesions. NEIL1 interacts with essential replication proteins such as the ring-shaped homotrimeric proliferating cellular nuclear antigen (PCNA). We isolated a complex formed between NEIL1 and PCNA (±DNA) using size exclusion chromatography (SEC). This interaction was confirmed using native gel electrophoresis and mass spectrometry. Stokes radii measured by SEC hinted that PCNA in complex with NEIL1 (±DNA) was no longer a trimer. Height measurements and images obtained by atomic force microscopy also demonstrated the dissociation of the PCNA homotrimer in the presence of NEIL1 and DNA, while small-angle X-ray scattering analysis confirmed the NEIL1 mediated PCNA trimer dissociation and formation of a 1:1:1 NEIL1-DNA-PCNA(monomer) complex. Furthermore, ab initio shape reconstruction provides insights into the solution structure of this previously unreported complex. Together, these data point to a potential mechanistic switch between replication and BER.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
The C-terminal tail of the NEIL1 DNA glycosylase interacts with the human mitochondrial single-stranded DNA binding protein.DNA Repair (Amst). 2018 May;65:11-19. doi: 10.1016/j.dnarep.2018.02.012. Epub 2018 Mar 6. DNA Repair (Amst). 2018. PMID: 29522991 Free PMC article.
-
Interaction of the human DNA glycosylase NEIL1 with proliferating cell nuclear antigen. The potential for replication-associated repair of oxidized bases in mammalian genomes.J Biol Chem. 2008 Feb 8;283(6):3130-3140. doi: 10.1074/jbc.M709186200. Epub 2007 Nov 21. J Biol Chem. 2008. PMID: 18032376
-
Physical and functional interaction between human oxidized base-specific DNA glycosylase NEIL1 and flap endonuclease 1.J Biol Chem. 2008 Oct 3;283(40):27028-37. doi: 10.1074/jbc.M802712200. Epub 2008 Jul 28. J Biol Chem. 2008. PMID: 18662981 Free PMC article.
-
New paradigms in the repair of oxidative damage in human genome: mechanisms ensuring repair of mutagenic base lesions during replication and involvement of accessory proteins.Cell Mol Life Sci. 2015 May;72(9):1679-98. doi: 10.1007/s00018-014-1820-z. Epub 2015 Jan 10. Cell Mol Life Sci. 2015. PMID: 25575562 Free PMC article. Review.
-
Decoding the Role of NEIL1 Gene in DNA Repair and Lifespan: A Literature Review with Bioinformatics Analysis.Adv Biol (Weinh). 2024 Nov;8(11):e2300708. doi: 10.1002/adbi.202300708. Epub 2024 Aug 20. Adv Biol (Weinh). 2024. PMID: 39164210 Review.
Cited by
-
Noncatalytic Domains in DNA Glycosylases.Int J Mol Sci. 2022 Jun 30;23(13):7286. doi: 10.3390/ijms23137286. Int J Mol Sci. 2022. PMID: 35806289 Free PMC article. Review.
-
The C-terminal tail of the NEIL1 DNA glycosylase interacts with the human mitochondrial single-stranded DNA binding protein.DNA Repair (Amst). 2018 May;65:11-19. doi: 10.1016/j.dnarep.2018.02.012. Epub 2018 Mar 6. DNA Repair (Amst). 2018. PMID: 29522991 Free PMC article.
-
Reconstruction of 3D density from solution scattering.Methods Enzymol. 2023;678:145-192. doi: 10.1016/bs.mie.2022.09.018. Epub 2022 Nov 3. Methods Enzymol. 2023. PMID: 36641207 Free PMC article.
-
Pre-Replicative Repair of Oxidized Bases Maintains Fidelity in Mammalian Genomes: The Cowcatcher Role of NEIL1 DNA Glycosylase.Genes (Basel). 2017 Jun 30;8(7):175. doi: 10.3390/genes8070175. Genes (Basel). 2017. PMID: 28665322 Free PMC article.
-
Recognition of DNA adducts by edited and unedited forms of DNA glycosylase NEIL1.DNA Repair (Amst). 2020 Jan;85:102741. doi: 10.1016/j.dnarep.2019.102741. Epub 2019 Nov 2. DNA Repair (Amst). 2020. PMID: 31733589 Free PMC article.
References
-
- De Bont R., van Larebeke N.. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis. 2004; 19:169–185. - PubMed
-
- Cooke M.S., Evans M.D., Dizdaroglu M., Lunec J.. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003; 17:1195–1214. - PubMed
-
- Cadet J., Douki T., Ravanat J.L.. Measurement of oxidatively generated base damage in cellular DNA. Mutat. Res. 2011; 711:3–12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

