Stabilization of Nucleotide Binding Domain Dimers Rescues ABCC6 Mutants Associated with Pseudoxanthoma Elasticum
- PMID: 27994049
- PMCID: PMC5290935
- DOI: 10.1074/jbc.M116.759811
Stabilization of Nucleotide Binding Domain Dimers Rescues ABCC6 Mutants Associated with Pseudoxanthoma Elasticum
Abstract
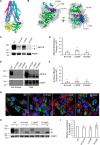
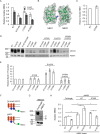
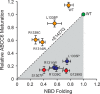
ABC transporters are polytopic membrane proteins that utilize ATP binding and hydrolysis to facilitate transport across biological membranes. Forty-eight human ABC transporters have been identified in the genome, and the majority of these are linked to heritable disease. Mutations in the ABCC6 (ATP binding cassette transporter C6) ABC transporter are associated with pseudoxanthoma elasticum, a disease of altered elastic properties in multiple tissues. Although ∼200 mutations have been identified in pseudoxanthoma elasticum patients, the underlying structural defects associated with the majority of these are poorly understood. To evaluate the structural consequences of these missense mutations, a combination of biophysical and cell biological approaches were applied to evaluate the local and global folding and assembly of the ABCC6 protein. Structural and bioinformatic analyses suggested that a cluster of mutations, representing roughly 20% of the patient population with identified missense mutations, are located in the interface between the transmembrane domain and the C-terminal nucleotide binding domain. Biochemical and cell biological analyses demonstrate these mutations influence multiple steps in the biosynthetic pathway, minimally altering local domain structure but adversely impacting ABCC6 assembly and trafficking. The differential impacts on local and global protein structure are consistent with hierarchical folding and assembly of ABCC6. Stabilization of specific domain-domain interactions via targeted amino acid substitution in the catalytic site of the C-terminal nucleotide binding domain restored proper protein trafficking and cell surface localization of multiple biosynthetic mutants. This rescue provides a specific mechanism by which chemical chaperones could be developed for the correction of ABCC6 biosynthetic defects.
Keywords: ABC transporter; abcc6; biosynthesis; calcification; protein stability; protein structure; pseudoxanthoma elasticum.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures


References
-
- Le Saux O., Urban Z., Tschuch C., Csiszar K., Bacchelli B., Quaglino D., Pasquali-Ronchetti I., Pope F. M., Richards A., Terry S., Bercovitch L., de Paepe A., and Boyd C. D. (2000) Mutations in a gene encoding an ABC transporter cause pseudoxanthoma elasticum. Nat. Genet. 25, 223–227 - PubMed
-
- Goodman R. M., Smith E. W., Paton D., Bergman R. A., Siegel C. L., Ottesen O. E., Shelley W. M., Pusch A. L., and McKusick V. A. (1963) Pseudoxanthoma elasticum: a clinical and histopathological study. Medicine 42, 297–334 - PubMed
-
- Jansen R. S., Duijst S., Mahakena S., Sommer D., Szeri F., Váradi A., Plomp A., Bergen A. A., Oude Elferink R. P., Borst P., and van de Wetering K. (2014) ABCC6-mediated ATP secretion by the liver is the main source of the mineralization inhibitor inorganic pyrophosphate in the systemic circulation-brief report. Arterioscler. Thromb. Vasc. Biol. 34, 1985–1989 - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

