Trp-Asp (WD) Repeat Domain 1 Is Essential for Mouse Peri-implantation Development and Regulates Cofilin Phosphorylation
- PMID: 27994054
- PMCID: PMC5270486
- DOI: 10.1074/jbc.M116.759886
Trp-Asp (WD) Repeat Domain 1 Is Essential for Mouse Peri-implantation Development and Regulates Cofilin Phosphorylation
Abstract
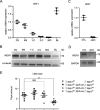
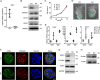
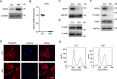
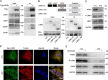
Trp-Asp (WD) repeat domain 1 (WDR1) is a highly conserved actin-binding protein across all eukaryotes and is involved in numerous actin-based processes by accelerating Cofilin severing actin filament. However, the function and the mechanism of WDR1 in mammalian early development are still largely unclear. We now report that WDR1 is essential for mouse peri-implantation development and regulates Cofilin phosphorylation in mouse cells. The disruption of maternal WDR1 does not obviously affect ovulation and female fertility. However, depletion of zygotic WDR1 results in embryonic lethality at the peri-implantation stage. In WDR1 knock-out cells, we found that WDR1 regulates Cofilin phosphorylation. Interestingly, WDR1 is overdosed to regulate Cofilin phosphorylation in mouse cells. Furthermore, we showed that WDR1 interacts with Lim domain kinase 1 (LIMK1), a well known phosphorylation kinase of Cofilin. Altogether, our results provide new insights into the role and mechanism of WDR1 in physiological conditions.
Keywords: Limk1; WDR1; actin; cofilin; development; embryonic development; mouse genetics; phosphorylation.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



Similar articles
-
Functions of actin-interacting protein 1 (AIP1)/WD repeat protein 1 (WDR1) in actin filament dynamics and cytoskeletal regulation.Biochem Biophys Res Commun. 2018 Nov 25;506(2):315-322. doi: 10.1016/j.bbrc.2017.10.096. Epub 2017 Oct 19. Biochem Biophys Res Commun. 2018. PMID: 29056508 Free PMC article. Review.
-
WDR1-regulated actin dynamics is required for outflow tract and right ventricle development.Dev Biol. 2018 Jun 15;438(2):124-137. doi: 10.1016/j.ydbio.2018.04.004. Epub 2018 Apr 11. Dev Biol. 2018. PMID: 29654745
-
Mutations in the cofilin partner Aip1/Wdr1 cause autoinflammatory disease and macrothrombocytopenia.Blood. 2007 Oct 1;110(7):2371-80. doi: 10.1182/blood-2006-10-055087. Epub 2007 May 21. Blood. 2007. PMID: 17515402 Free PMC article.
-
Involvement of LIMK1/2 in actin assembly during mouse embryo development.Cell Cycle. 2018;17(11):1381-1389. doi: 10.1080/15384101.2018.1482138. Epub 2018 Jul 25. Cell Cycle. 2018. PMID: 29943641 Free PMC article.
-
Regulation and signaling of the LIM domain kinases.Bioessays. 2025 Jan;47(1):e2400184. doi: 10.1002/bies.202400184. Epub 2024 Oct 3. Bioessays. 2025. PMID: 39361252 Review.
Cited by
-
Identification of potential therapeutic targets of deer antler extract on bone regulation based on serum proteomic analysis.Mol Biol Rep. 2019 Oct;46(5):4861-4872. doi: 10.1007/s11033-019-04934-0. Epub 2019 Jul 8. Mol Biol Rep. 2019. PMID: 31286391
-
Functions of actin-interacting protein 1 (AIP1)/WD repeat protein 1 (WDR1) in actin filament dynamics and cytoskeletal regulation.Biochem Biophys Res Commun. 2018 Nov 25;506(2):315-322. doi: 10.1016/j.bbrc.2017.10.096. Epub 2017 Oct 19. Biochem Biophys Res Commun. 2018. PMID: 29056508 Free PMC article. Review.
-
Coronin 1A depletion restores the nuclear stability and viability of Aip1/Wdr1-deficient neutrophils.J Cell Biol. 2019 Oct 7;218(10):3258-3271. doi: 10.1083/jcb.201901024. Epub 2019 Aug 30. J Cell Biol. 2019. PMID: 31471458 Free PMC article.
-
Network inference with Granger causality ensembles on single-cell transcriptomics.Cell Rep. 2022 Feb 8;38(6):110333. doi: 10.1016/j.celrep.2022.110333. Cell Rep. 2022. PMID: 35139376 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

