Intraclonal Genome Stability of the Metallo-β-lactamase SPM-1-producing Pseudomonas aeruginosa ST277, an Endemic Clone Disseminated in Brazilian Hospitals
- PMID: 27994579
- PMCID: PMC5136561
- DOI: 10.3389/fmicb.2016.01946
Intraclonal Genome Stability of the Metallo-β-lactamase SPM-1-producing Pseudomonas aeruginosa ST277, an Endemic Clone Disseminated in Brazilian Hospitals
Abstract
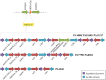
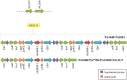
Carbapenems represent the mainstay therapy for the treatment of serious P. aeruginosa infections. However, the emergence of carbapenem resistance has jeopardized the clinical use of this important class of compounds. The production of SPM-1 metallo-β-lactamase has been the most common mechanism of carbapenem resistance identified in P. aeruginosa isolated from Brazilian medical centers. Interestingly, a single SPM-1-producing P. aeruginosa clone belonging to the ST277 has been widely spread within the Brazilian territory. In the current study, we performed a next-generation sequencing of six SPM-1-producing P. aeruginosa ST277 isolates. The core genome contains 5899 coding genes relative to the reference strain P. aeruginosa PAO1. A total of 26 genomic islands were detected in these isolates. We identified remarkable elements inside these genomic islands, such as copies of the blaSPM-1 gene conferring resistance to carbapenems and a type I-C CRISPR-Cas system, which is involved in protection of the chromosome against foreign DNA. In addition, we identified single nucleotide polymorphisms causing amino acid changes in antimicrobial resistance and virulence-related genes. Together, these factors could contribute to the marked resistance and persistence of the SPM-1-producing P. aeruginosa ST277 clone. A comparison of the SPM-1-producing P. aeruginosa ST277 genomes showed that their core genome has a high level nucleotide similarity and synteny conservation. The variability observed was mainly due to acquisition of genomic islands carrying several antibiotic resistance genes.
Keywords: Gram-negative bacilli; antimicrobial resistance; carbapenemase; comparative genomics; drug resistance; pathogenic bacteria.
Figures







Similar articles
-
Full characterization of the integrative and conjugative element carrying the metallo-β-lactamase bla SPM-1 and bicyclomycin bcr1 resistance genes found in the pandemic Pseudomonas aeruginosa clone SP/ST277.J Antimicrob Chemother. 2015 Sep;70(9):2547-50. doi: 10.1093/jac/dkv152. Epub 2015 Jun 20. J Antimicrob Chemother. 2015. PMID: 26093374
-
Exploring the success of Brazilian endemic clone Pseudomonas aeruginosa ST277 and its association with the CRISPR-Cas system type I-C.BMC Genomics. 2020 Mar 23;21(1):255. doi: 10.1186/s12864-020-6650-9. BMC Genomics. 2020. PMID: 32293244 Free PMC article.
-
SPM-1-producing Pseudomonas aeruginosa ST277 clone recovered from microbiota of migratory birds.Diagn Microbiol Infect Dis. 2018 Mar;90(3):221-227. doi: 10.1016/j.diagmicrobio.2017.11.003. Epub 2017 Nov 10. Diagn Microbiol Infect Dis. 2018. PMID: 29224710
-
Epidemiology and Characteristics of Metallo-β-Lactamase-Producing Pseudomonas aeruginosa.Infect Chemother. 2015 Jun;47(2):81-97. doi: 10.3947/ic.2015.47.2.81. Epub 2015 Jun 30. Infect Chemother. 2015. PMID: 26157586 Free PMC article. Review.
-
Mobile Carbapenemase Genes in Pseudomonas aeruginosa.Front Microbiol. 2021 Feb 18;12:614058. doi: 10.3389/fmicb.2021.614058. eCollection 2021. Front Microbiol. 2021. PMID: 33679638 Free PMC article. Review.
Cited by
-
Metallo-β-lactamases in the Age of Multidrug Resistance: From Structure and Mechanism to Evolution, Dissemination, and Inhibitor Design.Chem Rev. 2021 Jul 14;121(13):7957-8094. doi: 10.1021/acs.chemrev.1c00138. Epub 2021 Jun 15. Chem Rev. 2021. PMID: 34129337 Free PMC article. Review.
-
Geographic and Temporal Patterns of Antimicrobial Resistance in Pseudomonas aeruginosa Over 20 Years From the SENTRY Antimicrobial Surveillance Program, 1997-2016.Open Forum Infect Dis. 2019 Mar 15;6(Suppl 1):S63-S68. doi: 10.1093/ofid/ofy343. eCollection 2019 Mar. Open Forum Infect Dis. 2019. PMID: 30895216 Free PMC article.
-
Diversity and Distribution of Resistance Markers in Pseudomonas aeruginosa International High-Risk Clones.Microorganisms. 2021 Feb 12;9(2):359. doi: 10.3390/microorganisms9020359. Microorganisms. 2021. PMID: 33673029 Free PMC article. Review.
-
Comprehensive genome data analysis establishes a triple whammy of carbapenemases, ICEs and multiple clinically relevant bacteria.Microb Genom. 2020 Oct;6(10):mgen000424. doi: 10.1099/mgen.0.000424. Microb Genom. 2020. PMID: 32841111 Free PMC article.
-
Phenotyping and Genotyping Evaluation of E. coli Produces Carbapenemase Isolated from Cancer Patients in Al-Basrah, Iraq.Arch Razi Inst. 2023 Jun 30;78(3):823-829. doi: 10.22092/ARI.2022.359869.2493. eCollection 2023 Jun. Arch Razi Inst. 2023. PMID: 38028834 Free PMC article.
References
-
- Al-Nayyef H., Guyeux C., Petitjean M., Hocquet D., Bahi J. (2015). Relation between insertion sequences and genome rearrangements in Pseudomonas aeruginosa, in Bioinformatics and Biomedical Engineering, eds Ortuño F., Rojas I. (Granada: Springer International Publishing; ), 426–437.
LinkOut - more resources
Full Text Sources
Other Literature Sources

