TRIAD3/RNF216 mutations associated with Gordon Holmes syndrome lead to synaptic and cognitive impairments via Arc misregulation
- PMID: 27995769
- PMCID: PMC5334534
- DOI: 10.1111/acel.12551
TRIAD3/RNF216 mutations associated with Gordon Holmes syndrome lead to synaptic and cognitive impairments via Arc misregulation
Abstract
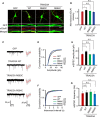
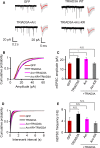
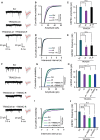
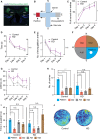
Multiple loss-of-function mutations in TRIAD3 (a.k.a. RNF216) have recently been identified in patients suffering from Gordon Holmes syndrome (GHS), characterized by cognitive decline, dementia, and movement disorders. TRIAD3A is an E3 ubiquitin ligase that recognizes and facilitates the ubiquitination of its target for degradation by the ubiquitin-proteasome system (UPS). Here, we demonstrate that two of these missense substitutions in TRIAD3 (R660C and R694C) could not regulate the degradation of their neuronal target, activity-regulated cytoskeletal-associated protein (Arc/Arg 3.1), whose expression is critical for synaptic plasticity and memory. The synaptic deficits due to the loss of endogenous TRIAD3A could not be rescued by TRIAD3A harboring GHS-associated missense mutations. Moreover, we demonstrate that the loss of endogenous TRIAD3A in the mouse hippocampal CA1 region led to deficits in spatial learning and memory. Finally, we show that these missense mutations abolished the interaction of TRIAD3A with Arc, disrupting Arc ubiquitination, and consequently Arc degradation. Our current findings of Arc misregulation by TRIAD3A variants suggest that loss-of-function mutations in TRIAD3A may contribute to dementia observed in patients with GHS driven by dysfunctional UPS components, leading to cognitive impairments through the synaptic protein Arc.
Keywords: Gordon Holmes syndrome; TRIAD3; activity-regulated cytoskeletal protein (Arc/Arg 3.1); behavior; dementia; learning and memory; synapse; ubiquitination.
© 2016 The Authors. Aging Cell published by the Anatomical Society and John Wiley & Sons Ltd.
Figures

References
-
- Abs R, Van Vleymen E, Parizel PM, Van Acker K, Martin M, Martin JJ (1990) Congenital cerebellar hypoplasia and hypogonadotropic hypogonadism. J. Neurol. Sci. 98, 259–265. - PubMed
-
- Chen D, Li X, Zhai Z, Shu HB (2002) A novel zinc finger protein interacts with receptor‐interacting protein (RIP) and inhibits tumor necrosis factor (TNF)‐ and IL1‐induced NF‐kappa B activation. J. Biol. Chem. 277, 15985–15991. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

