RNA activation of haploinsufficient Foxg1 gene in murine neocortex
- PMID: 27995975
- PMCID: PMC5172352
- DOI: 10.1038/srep39311
RNA activation of haploinsufficient Foxg1 gene in murine neocortex
Abstract
More than one hundred distinct gene hemizygosities are specifically linked to epilepsy, mental retardation, autism, schizophrenia and neuro-degeneration. Radical repair of these gene deficits via genome engineering is hardly feasible. The same applies to therapeutic stimulation of the spared allele by artificial transactivators. Small activating RNAs (saRNAs) offer an alternative, appealing approach. As a proof-of-principle, here we tested this approach on the Rett syndrome-linked, haploinsufficient, Foxg1 brain patterning gene. We selected a set of artificial small activating RNAs (saRNAs) upregulating it in neocortical precursors and their derivatives. Expression of these effectors achieved a robust biological outcome. saRNA-driven activation (RNAa) was limited to neural cells which normally express Foxg1 and did not hide endogenous gene tuning. saRNAs recognized target chromatin through a ncRNA stemming from it. Gene upregulation required Ago1 and was associated to RNApolII enrichment throughout the Foxg1 locus. Finally, saRNA delivery to murine neonatal brain replicated Foxg1-RNAa in vivo.
Conflict of interest statement
C.F., E.G., Q.S. and A.M. do not declare any conflicts. G.G. is a co-founder of Voyager Therapeutics specialized in rAAV-based gene therapy, and holds equity in the company. Moreover, G.G. is also an inventor on patents with potential royalties licensed to Voyager Therapeutics and other biopharmaceutical companies.
Figures

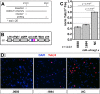


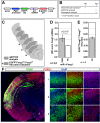
References
-
- Carvill G. L. & Mefford H. C. Microdeletion syndromes. Current Opinion in Genetics & Development 23, 232–239 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

