Three-Dimensional Structure Determination of Surface Sites
- PMID: 27997167
- PMCID: PMC5719466
- DOI: 10.1021/jacs.6b10894
Three-Dimensional Structure Determination of Surface Sites
Abstract
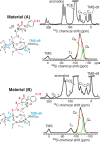
The spatial arrangement of atoms is directly linked to chemical function. A fundamental challenge in surface chemistry and catalysis relates to the determination of three-dimensional structures with atomic-level precision. Here we determine the three-dimensional structure of an organometallic complex on an amorphous silica surface using solid-state NMR measurements, enabled through a dynamic nuclear polarization surface enhanced NMR spectroscopy approach that induces a 200-fold increase in the NMR sensitivity for the surface species. The result, in combination with EXAFS, is a detailed structure for the surface complex determined with a precision of 0.7 Å. We observe a single well-defined conformation that is folded toward the surface in such a way as to include an interaction between the platinum metal center and the surface oxygen atoms.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

