Identification and Characterisation of a Novel Protein FIP-sch3 from Stachybotrys chartarum
- PMID: 27997578
- PMCID: PMC5173029
- DOI: 10.1371/journal.pone.0168436
Identification and Characterisation of a Novel Protein FIP-sch3 from Stachybotrys chartarum
Abstract
In this study, a novel FIP named FIP-sch3 has been identified and characterised. FIP-sch3 was identified in the ascomycete Stachybotrys chartarum, making it the second FIP to be identified outside the order of Basidiomycota. Recombinant FIP-sch3 (rFIP-shc3) was produced in Escherichia coli and purified using GST-affinity magnetic beads. The bioactive characteristics of FIP-sch3 were compared to those of well-known FIPs LZ-8 from Ganoderma lucidum and FIP-fve from Flammulina velutipes, which were produced and purified using the same method. The purified rFIP-sch3 exhibited a broad spectrum of anti-tumour activity in several types of tumour cells but had no cytotoxicity in normal human embryonic kidney 293 cells. Assays that were implemented to study these properties indicated that rFIP-sch3 significantly suppressed cell proliferation, induced apoptosis and inhibited cell migration in human lung adenocarcinoma A549 cells. The anti-tumour effects of rFIP-sch3 in A549 cells were comparable to those of rLZ-8, but they were significantly greater than those of rFIP-fve. Molecular assays that were built on real-time PCR further revealed potential mechanisms related to apoptosis and migration and that underlie phenotypic effects. These results indicate that FIP-shc3 has a unique anti-tumour bioactive profile, as do other FIPs, which provide a foundation for further studies on anti-tumour mechanisms. Importantly, this study also had convenient access to FIP-sch3 with potential human therapeutic applications.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





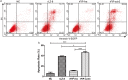


References
-
- Wang XF, Su KQ, Bao TW, Cong WR, Chen YF, Li QZ, et al. Immunomodulatory effects of fungal proteins. Curr Top Nutraceutical Res. 2012;10:1–11.
-
- Paaventhan P, Joseph JS, Seow SV, Vaday S, Robinson H, Chua KY, et al. A 1.7Å structure of Fve, a member of the new fungal immunomodulatory protein family. J Mol Biol. 2003;332(2):461–70. - PubMed
-
- Huang L, Sun F, Liang C, He YX, Bao R, Liu L, et al. Crystal structure of LZ-8 from the medicinal fungus Ganoderma lucidum. Proteins. 2008;75(2):524–7. - PubMed
-
- Ko JL, Hsu CI, Lin RH, Kao CL, Lin JY. A new fungal immunomodulatory protein, FIP-fve isolated from the edible mushroom, Flammulina velutipes and its complete amino acid sequence. Eur J Biochem. 1995;228(2):244–9. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

