The C-Terminal SynMuv/DdDUF926 Domain Regulates the Function of the N-Terminal Domain of DdNKAP
- PMID: 27997579
- PMCID: PMC5173251
- DOI: 10.1371/journal.pone.0168617
The C-Terminal SynMuv/DdDUF926 Domain Regulates the Function of the N-Terminal Domain of DdNKAP
Abstract
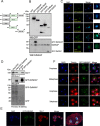
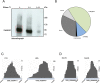
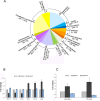
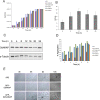
NKAP (NF-κB activating protein) is a highly conserved SR (serine/arginine-rich) protein involved in transcriptional control and splicing in mammals. We identified DdNKAP, the Dictyostelium discoideum ortholog of mammalian NKAP, as interacting partner of the nuclear envelope protein SUN-1. DdNKAP harbors a number of basic RDR/RDRS repeats in its N-terminal domain and the SynMuv/DUF926 domain at its C-terminus. We describe a novel and direct interaction between DdNKAP and Prp19 (Pre mRNA processing factor 19) which might be relevant for the observed DdNKAP ubiquitination. Genome wide analysis using cross-linking immunoprecipitation-high-throughput sequencing (CLIP-seq) revealed DdNKAP association with intergenic regions, exons, introns and non-coding RNAs. Ectopic expression of DdNKAP and its domains affects several developmental aspects like stream formation, aggregation, and chemotaxis. We conclude that DdNKAP is a multifunctional protein, which might influence Dictyostelium development through its interaction with RNA and RNA binding proteins. Mutants overexpressing full length DdNKAP and the N-terminal domain alone (DdN-NKAP) showed opposite phenotypes in development and opposite expression profiles of several genes and rRNAs. The observed interaction between DdN-NKAP and the DdDUF926 domain indicates that the DdDUF926 domain acts as negative regulator of the N-terminus.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Chen D, Li Z, Yang Q, Zhang J, Zhai Z, Shu HB. Identification of a nuclear protein that promotes NF-kappaB activation. Biochem Biophys Res Commun. 2003;310: 720–724. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

