Prediction of Possible Biomarkers and Novel Pathways Conferring Risk to Post-Traumatic Stress Disorder
- PMID: 27997584
- PMCID: PMC5172609
- DOI: 10.1371/journal.pone.0168404
Prediction of Possible Biomarkers and Novel Pathways Conferring Risk to Post-Traumatic Stress Disorder
Abstract
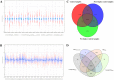
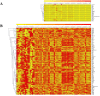
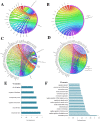
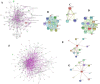
Post-traumatic stress disorder is one of the common mental ailments that is triggered by exposure to traumatic events. Till date, the molecular factors conferring risk to the development of PTSD is not well understood. In this study, we have conducted a meta-analysis followed by hierarchical clustering and functional enrichment, to uncover the potential molecular networks and critical genes which play an important role in PTSD. Two datasets of expression profiles from Peripheral Blood Mononuclear Cells from 62 control samples and 63 PTSD samples were included in our study. In PTSD samples of GSE860 dataset, we identified 26 genes informative when compared with Post-deploy PTSD condition and 58 genes informative when compared with Pre-deploy and Post-deploy PTSD of GSE63878 dataset. We conducted the meta-analysis using Fisher, roP, Stouffer, AW, SR, PR and RP methods in MetaDE package. Results from the rOP method of MetaDE package showed that among these genes, the following showed significant changes including, OR2B6, SOX21, MOBP, IL15, PTPRK, PPBPP2 and SEC14L5. Gene ontology analysis revealed enrichment of these significant PTSD-related genes for cell proliferation, DNA damage and repair (p-value ≤ 0.05). Furthermore, interaction network analysis was performed on these 7 significant genes. This analysis revealed highly connected functional interaction networks with two candidate genes, IL15 and SEC14L5 highly enriched in networks. Overall, from these results, we concluded that these genes can be recommended as some of the potential targets for PTSD.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Differential transcriptional response following glucocorticoid activation in cultured blood immune cells: a novel approach to PTSD biomarker development.Transl Psychiatry. 2019 Aug 21;9(1):201. doi: 10.1038/s41398-019-0539-x. Transl Psychiatry. 2019. PMID: 31434874 Free PMC article.
-
Transcriptome analysis reveals novel genes and immune networks dysregulated in veterans with PTSD.Brain Behav Immun. 2018 Nov;74:133-142. doi: 10.1016/j.bbi.2018.08.014. Epub 2018 Sep 4. Brain Behav Immun. 2018. PMID: 30189241
-
Gene networks specific for innate immunity define post-traumatic stress disorder.Mol Psychiatry. 2015 Dec;20(12):1538-45. doi: 10.1038/mp.2015.9. Epub 2015 Mar 10. Mol Psychiatry. 2015. PMID: 25754082 Free PMC article.
-
[Posttraumatic stress disorder (PTSD) as a consequence of the interaction between an individual genetic susceptibility, a traumatogenic event and a social context].Encephale. 2012 Oct;38(5):373-80. doi: 10.1016/j.encep.2011.12.003. Epub 2012 Jan 24. Encephale. 2012. PMID: 23062450 Review. French.
-
Genetic Advances in Post-traumatic Stress Disorder.Rev Colomb Psiquiatr (Engl Ed). 2018 Apr-Jun;47(2):108-118. doi: 10.1016/j.rcp.2016.12.001. Epub 2017 Jan 9. Rev Colomb Psiquiatr (Engl Ed). 2018. PMID: 29754704 Review. English, Spanish.
Cited by
-
Protein tyrosine phosphatase receptor type kappa (PTPRK) revisited: evolving insights into structure, function, and pathology.J Transl Med. 2025 May 12;23(1):534. doi: 10.1186/s12967-025-06496-1. J Transl Med. 2025. PMID: 40355891 Free PMC article. Review.
-
Computational analysis of deleterious single nucleotide polymorphisms in catechol O-Methyltransferase conferring risk to post-traumatic stress disorder.J Psychiatr Res. 2021 Jun;138:207-218. doi: 10.1016/j.jpsychires.2021.03.048. Epub 2021 Mar 31. J Psychiatr Res. 2021. PMID: 33865170 Free PMC article.
-
Epigenome-Wide Study of Posttraumatic Stress Disorder Symptom Severity in a Treatment-Seeking Adolescent Sample.J Trauma Stress. 2021 Jun;34(3):607-615. doi: 10.1002/jts.22655. Epub 2021 Feb 2. J Trauma Stress. 2021. PMID: 33529416 Free PMC article.
-
Neurotensin and Neurotensin Receptors in Stress-related Disorders: Pathophysiology & Novel Drug Targets.Curr Neuropharmacol. 2024;22(5):916-934. doi: 10.2174/1570159X21666230803101629. Curr Neuropharmacol. 2024. PMID: 37534788 Free PMC article. Review.
-
Disentangling the effects of PTSD from Gulf War Illness in male veterans via a systems-wide analysis of immune cell, cytokine, and symptom measures.Mil Med Res. 2024 Jan 2;11(1):2. doi: 10.1186/s40779-023-00505-4. Mil Med Res. 2024. PMID: 38167090 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

