Metagenomic profiling of gut microbial communities in both wild and artificially reared Bar-headed goose (Anser indicus)
- PMID: 27998035
- PMCID: PMC5387313
- DOI: 10.1002/mbo3.429
Metagenomic profiling of gut microbial communities in both wild and artificially reared Bar-headed goose (Anser indicus)
Abstract
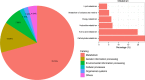
Bar-headed goose (Anser indicus), a species endemic to Asia, has become one of the most popular species in recent years for rare bird breeding industries in several provinces of China. There has been no information on the gut metagenome configuration in both wild and artificially reared Bar-headed geese, even though the importance of gut microbiome in vertebrate nutrient and energy metabolism, immune homeostasis and reproduction is widely acknowledged. In this study, metagenomic methods have been used to describe the microbial community structure and composition of functional genes associated with both wild and artificially reared Bar-headed goose. Taxonomic analyses revealed that Firmicutes, Proteobacteria, Actinobacteria and Bacteroidetes were the four most abundant phyla in the gut of Bar-headed geese. Bacteroidetes were significantly abundant in the artificially reared group compared to wild group. Through functional profiling, we found that artificially reared Bar-headed geese had higher bacterial gene content related to carbohydrate transport and metabolism, energy metabolism and coenzyme transport, and metabolism. A comprehensive gene catalog of Bar-headed geese metagenome was built, and the metabolism of carbohydrate, amino acid, nucleotide, and energy were found to be the four most abundant categories. These results create a baseline for future Bar-headed goose microbiology research, and make an original contribution to the artificial rearing of this bird.
Keywords: Bar-headed goose; gut microbiome; high-throughput sequencing; metagenomics; taxonomic analysis.
© 2016 The Authors. MicrobiologyOpen published by John Wiley & Sons Ltd.
Figures


References
-
- Andrews, S . (2012). FastQC: A Quality Control tool for High Throughput Sequence Data. Available from http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed Nov. 20, 2015.
-
- Backhed, F. , Ley, R. E. , Sonnenburg, J. L. , Peterson, D. A. , & Gordon, J. I. (2005). Host‐bacterial mutualism in the human intestine. Science, 307, 1915–1920. - PubMed
-
- Bishop, M. A. , Song, Y. L. , Canjue, Z. M. , & Gu, B. Y. (1997). Bar‐headed Geese Anser indicus wintering in South‐central Tibet. Wildfowl, 48, 118–126.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

