Methylome evolution in plants
- PMID: 27998290
- PMCID: PMC5175322
- DOI: 10.1186/s13059-016-1127-5
Methylome evolution in plants
Erratum in
-
Erratum to: Methylome Evolution in plants.Genome Biol. 2017 Feb 27;18(1):41. doi: 10.1186/s13059-017-1176-4. Genome Biol. 2017. PMID: 28241876 Free PMC article. No abstract available.
Abstract
Despite major progress in dissecting the molecular pathways that control DNA methylation patterns in plants, little is known about the mechanisms that shape plant methylomes over evolutionary time. Drawing on recent intra- and interspecific epigenomic studies, we show that methylome evolution over long timescales is largely a byproduct of genomic changes. By contrast, methylome evolution over short timescales appears to be driven mainly by spontaneous epimutational events. We argue that novel methods based on analyses of the methylation site frequency spectrum (mSFS) of natural populations can provide deeper insights into the evolutionary forces that act at each timescale.
Figures

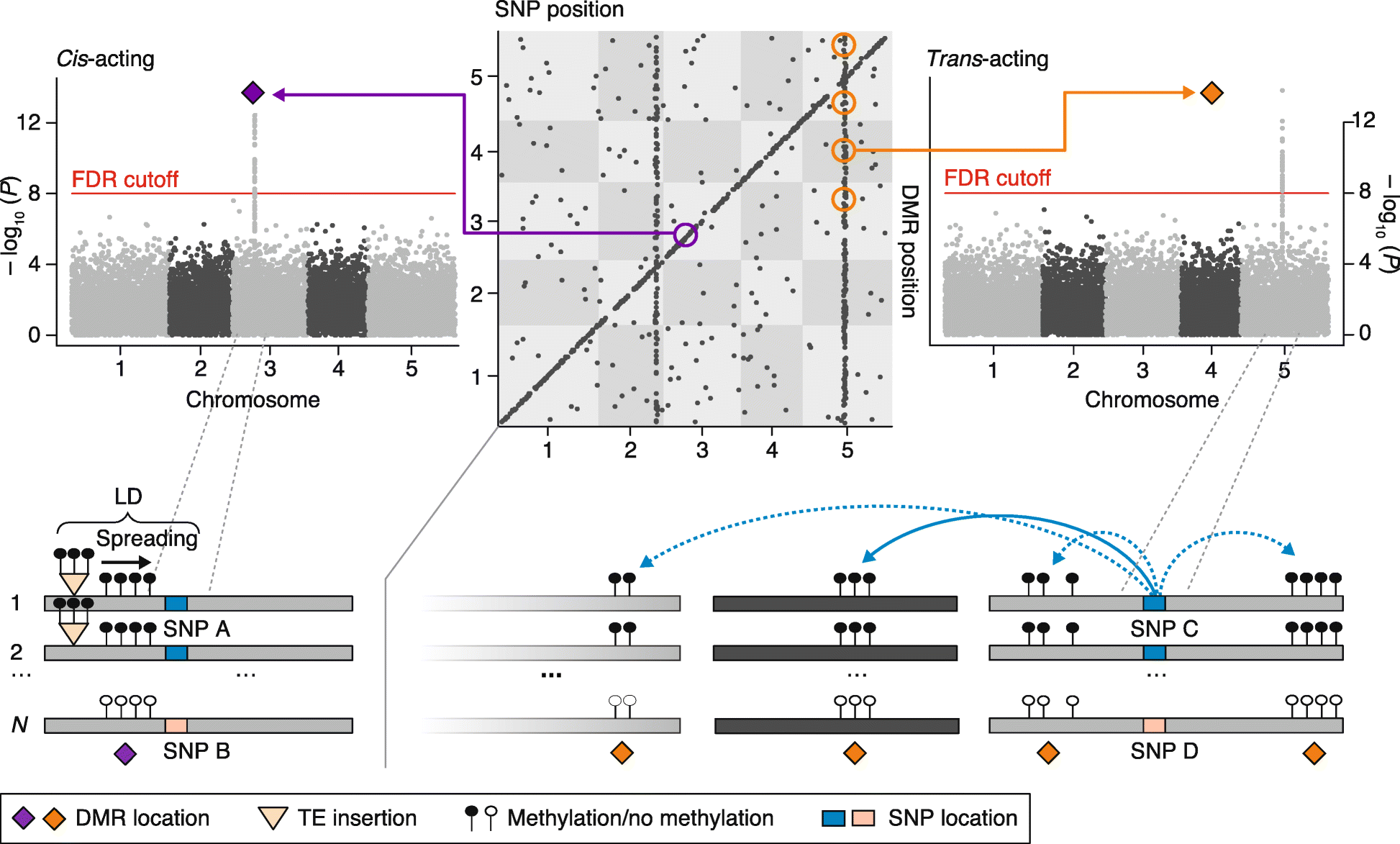


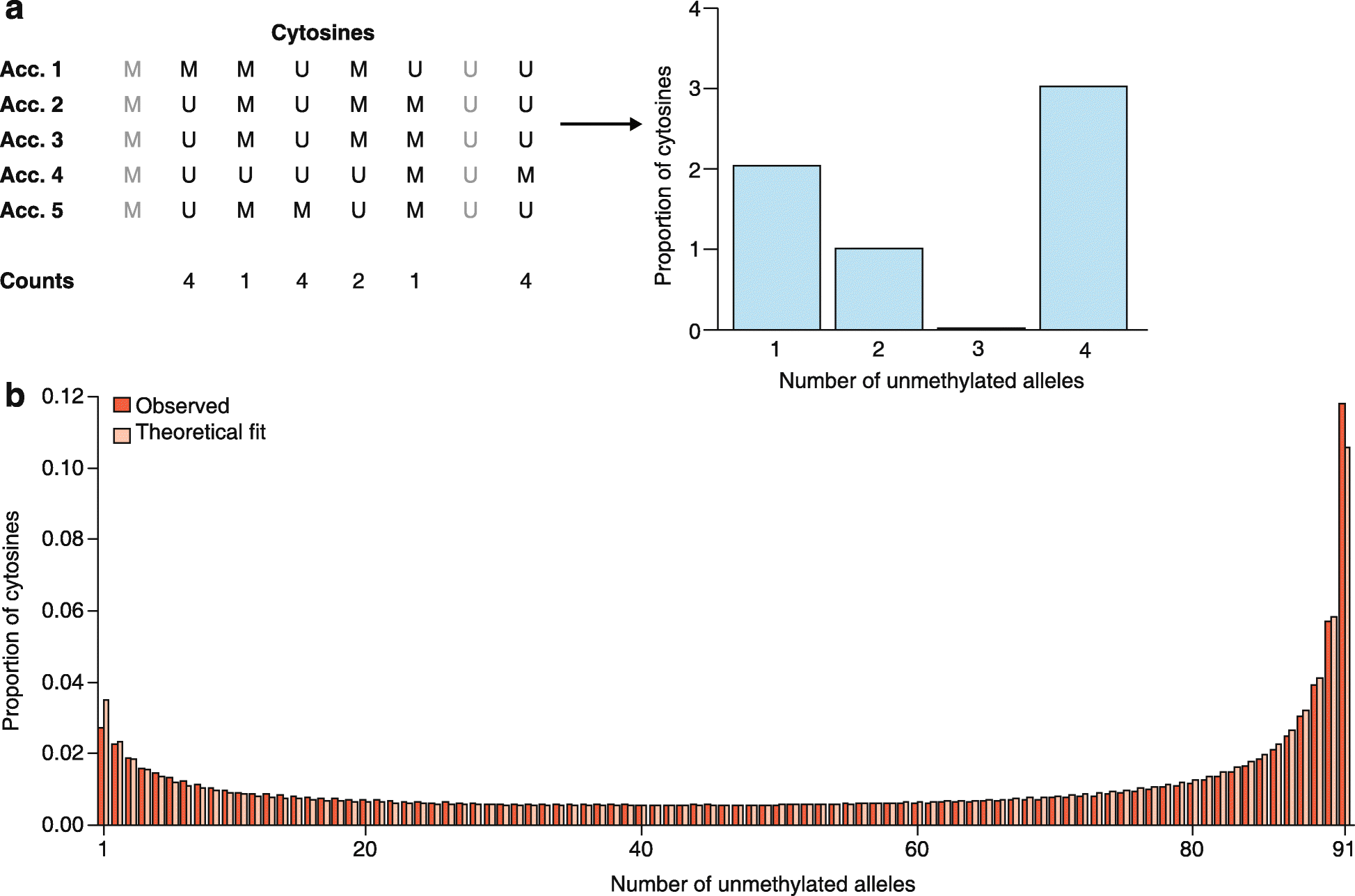
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

