Resilient microorganisms in dust samples of the International Space Station-survival of the adaptation specialists
- PMID: 27998314
- PMCID: PMC5175303
- DOI: 10.1186/s40168-016-0217-7
Resilient microorganisms in dust samples of the International Space Station-survival of the adaptation specialists
Abstract
Background: The International Space Station (ISS) represents a unique biotope for the human crew but also for introduced microorganisms. Microbes experience selective pressures such as microgravity, desiccation, poor nutrient-availability due to cleaning, and an increased radiation level. We hypothesized that the microbial community inside the ISS is modified by adapting to these stresses. For this reason, we analyzed 8-12 years old dust samples from Russian ISS modules with major focus on the long-time surviving portion of the microbial community. We consequently assessed the cultivable microbiota of these samples in order to analyze their extremotolerant potential against desiccation, heat-shock, and clinically relevant antibiotics. In addition, we studied the bacterial and archaeal communities from the stored Russian dust samples via molecular methods (next-generation sequencing, NGS) and compared our new data with previously derived information from the US American ISS dust microbiome.
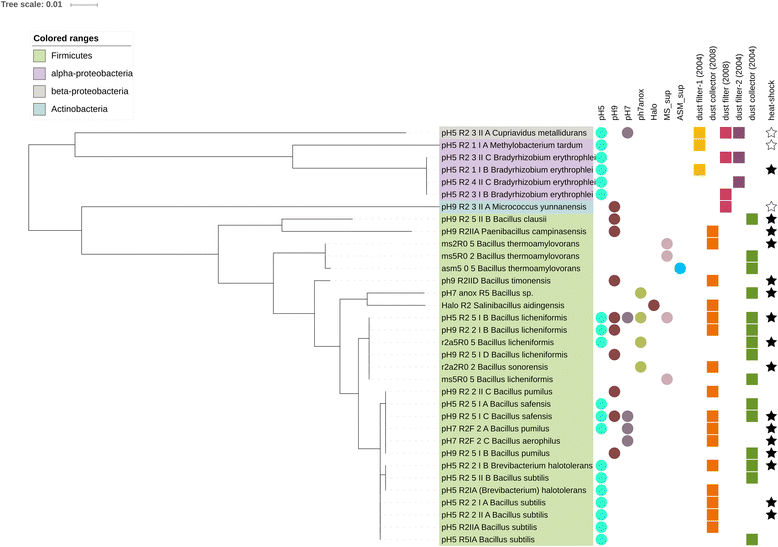
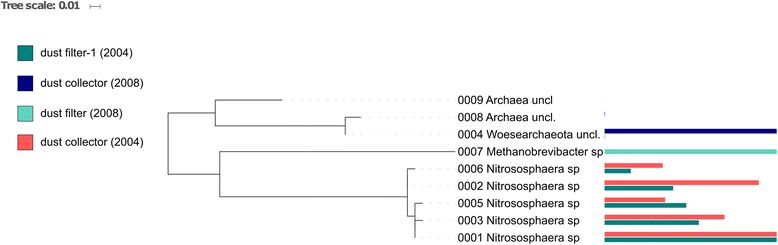
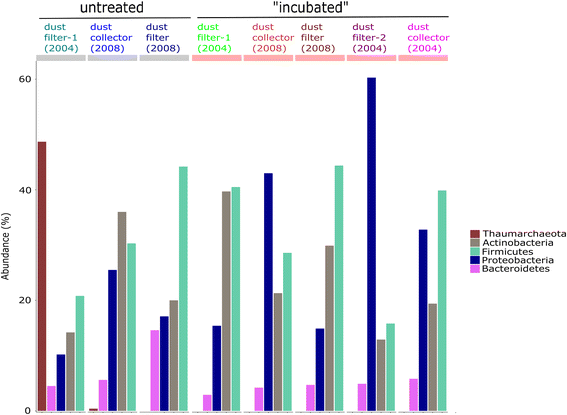
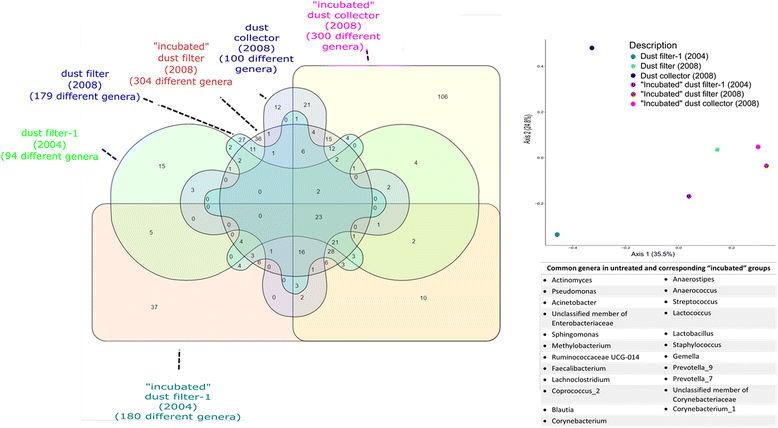
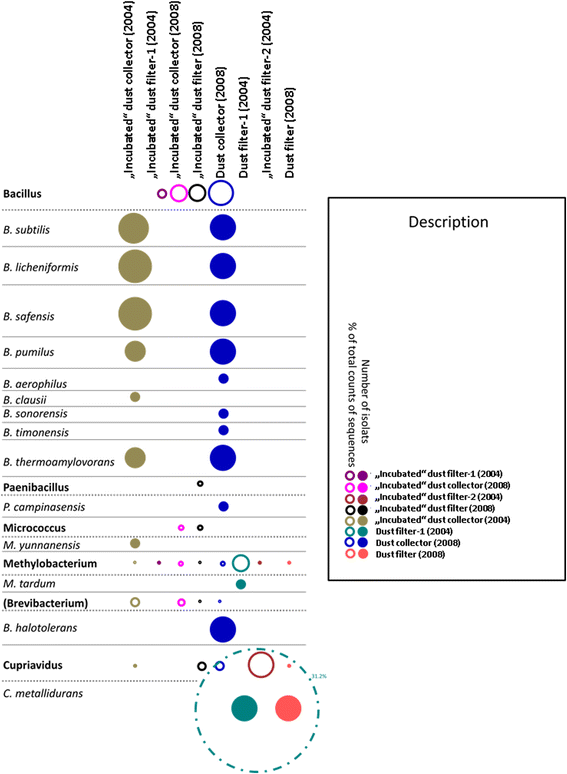
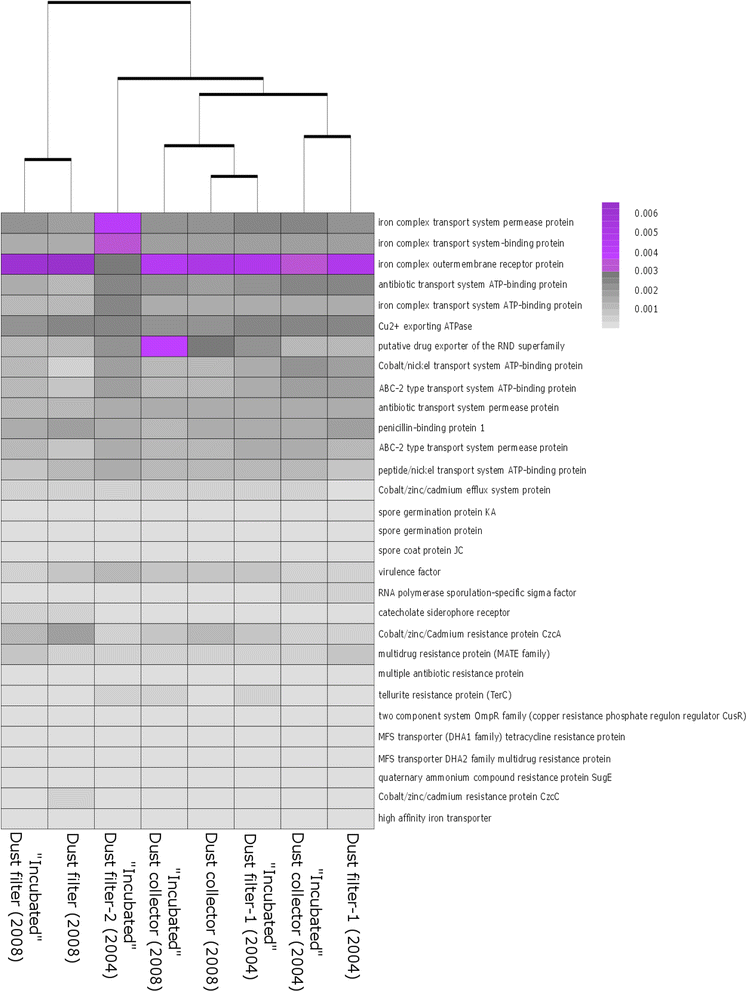
Results: We cultivated and identified in total 85 bacterial, non-pathogenic isolates (17 different species) and 1 fungal isolate from the 8-12 year old dust samples collected in the Russian segment of the ISS. Most of these isolates exhibited robust resistance against heat-shock and clinically relevant antibiotics. Microbial 16S rRNA gene and archaeal 16S rRNA gene targeting Next Generation Sequencing showed signatures of human-associated microorganisms (Corynebacterium, Staphylococcus, Coprococcus etc.), but also specifically adapted extremotolerant microorganisms. Besides bacteria, the detection of archaeal signatures in higher abundance was striking.
Conclusions: Our findings reveal (i) the occurrence of living, hardy microorganisms in archived Russian ISS dust samples, (ii) a profound resistance capacity of ISS microorganisms against environmental stresses, and (iii) the presence of archaeal signatures on board. In addition, we found indications that the microbial community in the Russian segment dust samples was different to recently reported US American ISS microbiota.
Keywords: Archaea; Confined habitat; Extremotolerant; International Space Station; Microbiome.
Figures
References
-
- Mora M, Mahnert A, Koskinen K, Pausan MR, Oberauner-Wappis L, Krause R, et al. Microorganisms in confined habitats: microbial monitoring and control of the International Space Station, cleanrooms, operating rooms and intensive care units. Front. Microbiol. 2016; 7:1573. Available from: http://dx.doi.org/10.3389/fmicb.2016.01573. Accessed 7 Dec 2016. - DOI - PMC - PubMed
-
- Moissl-Eichinger C, Cockell C, Rettberg P. Venturing into new realms? Microorganisms in space. FEMS Microbiol Rev. 2016;40(5):722–37. - PubMed
-
- Venkateswaran K, Vaishampayan P, Cisneros J, Pierson DL, Rogers SO, Perry J. International Space Station environmental microbiome- microbial inventories of ISS filter debris. Appl Microbiol Biotechnol. 2014: 98. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

