Xenolog classification
- PMID: 27998934
- PMCID: PMC5860392
- DOI: 10.1093/bioinformatics/btw686
Xenolog classification
Abstract
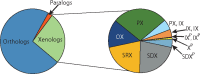
Motivation: Orthology analysis is a fundamental tool in comparative genomics. Sophisticated methods have been developed to distinguish between orthologs and paralogs and to classify paralogs into subtypes depending on the duplication mechanism and timing, relative to speciation. However, no comparable framework exists for xenologs: gene pairs whose history, since their divergence, includes a horizontal transfer. Further, the diversity of gene pairs that meet this broad definition calls for classification of xenologs with similar properties into subtypes.
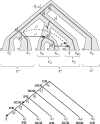
Results: We present a xenolog classification that uses phylogenetic reconciliation to assign each pair of genes to a class based on the event responsible for their divergence and the historical association between genes and species. Our classes distinguish between genes related through transfer alone and genes related through duplication and transfer. Further, they separate closely-related genes in distantly-related species from distantly-related genes in closely-related species. We present formal rules that assign gene pairs to specific xenolog classes, given a reconciled gene tree with an arbitrary number of duplications and transfers. These xenology classification rules have been implemented in software and tested on a collection of ∼13 000 prokaryotic gene families. In addition, we present a case study demonstrating the connection between xenolog classification and gene function prediction.
Availability and implementation: The xenolog classification rules have been implemented in N otung 2.9, a freely available phylogenetic reconciliation software package. http://www.cs.cmu.edu/~durand/Notung . Gene trees are available at http://dx.doi.org/10.7488/ds/1503 .
Contact: durand@cmu.edu.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author 2016. Published by Oxford University Press.
Figures





References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

